Differentiating Benign from Malignant Thyroid Tumors by Kinase Activity Profiling and Dabrafenib BRAF V600E Targeting
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Material
2.2. Nucleic Acid Isolation, DNA Variant, and Fusion Analysis
2.3. Materials
2.4. Preparation of Lysates
2.5. Kinase Activity Assays
2.6. Data Quantification
2.7. Classification of Benign and Malignant Samples
2.8. Statistical Analysis
2.9. Upstream Kinase Analysis
2.10. Network Analysis
3. Results
3.1. Patient Characteristics
3.2. Kinase Activity Profiling Classifies Benign and Malignant Thyroid Tumors
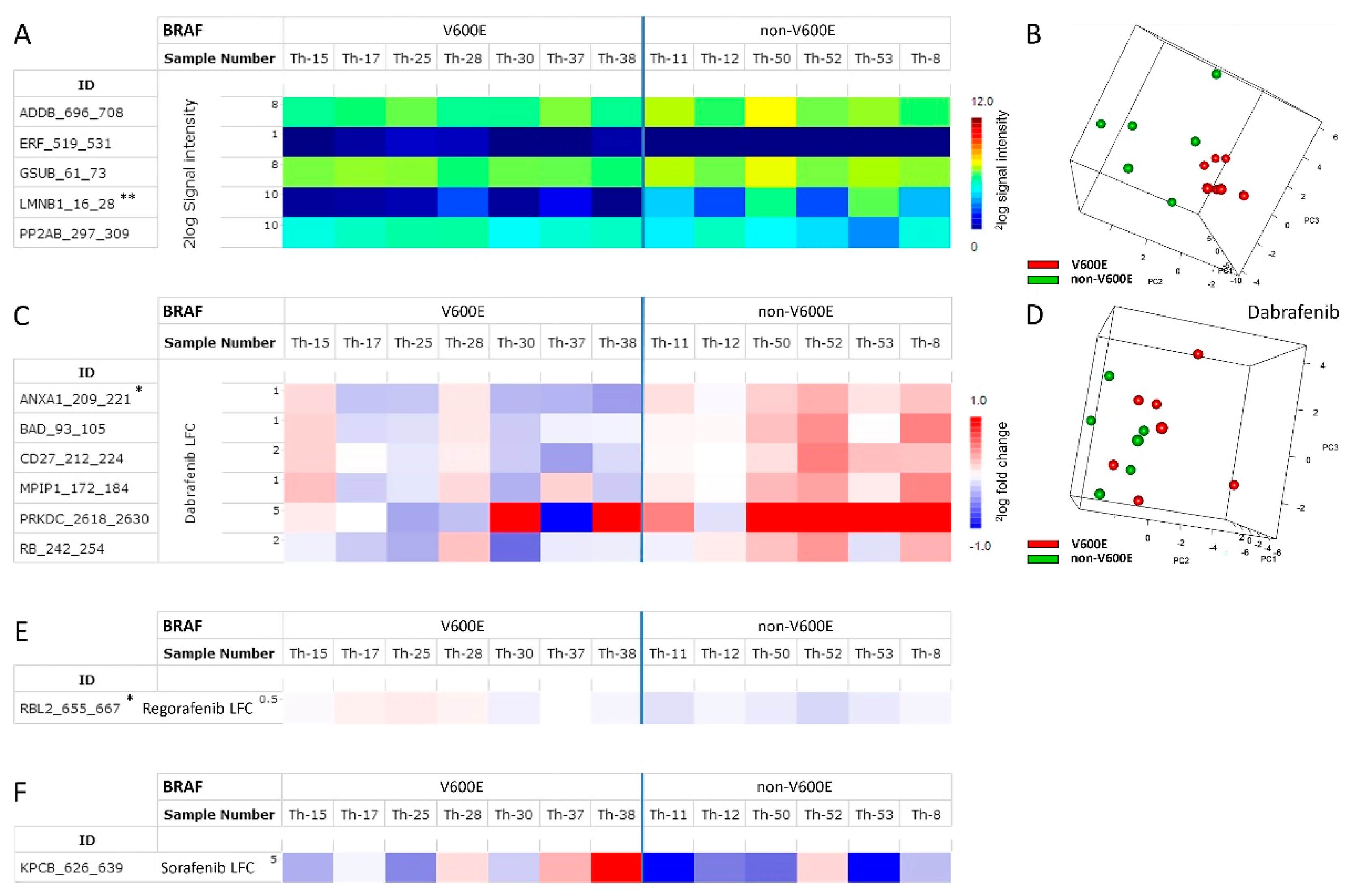
3.3. Molecular Interpretation of Differences between Benign and Malignant Samples
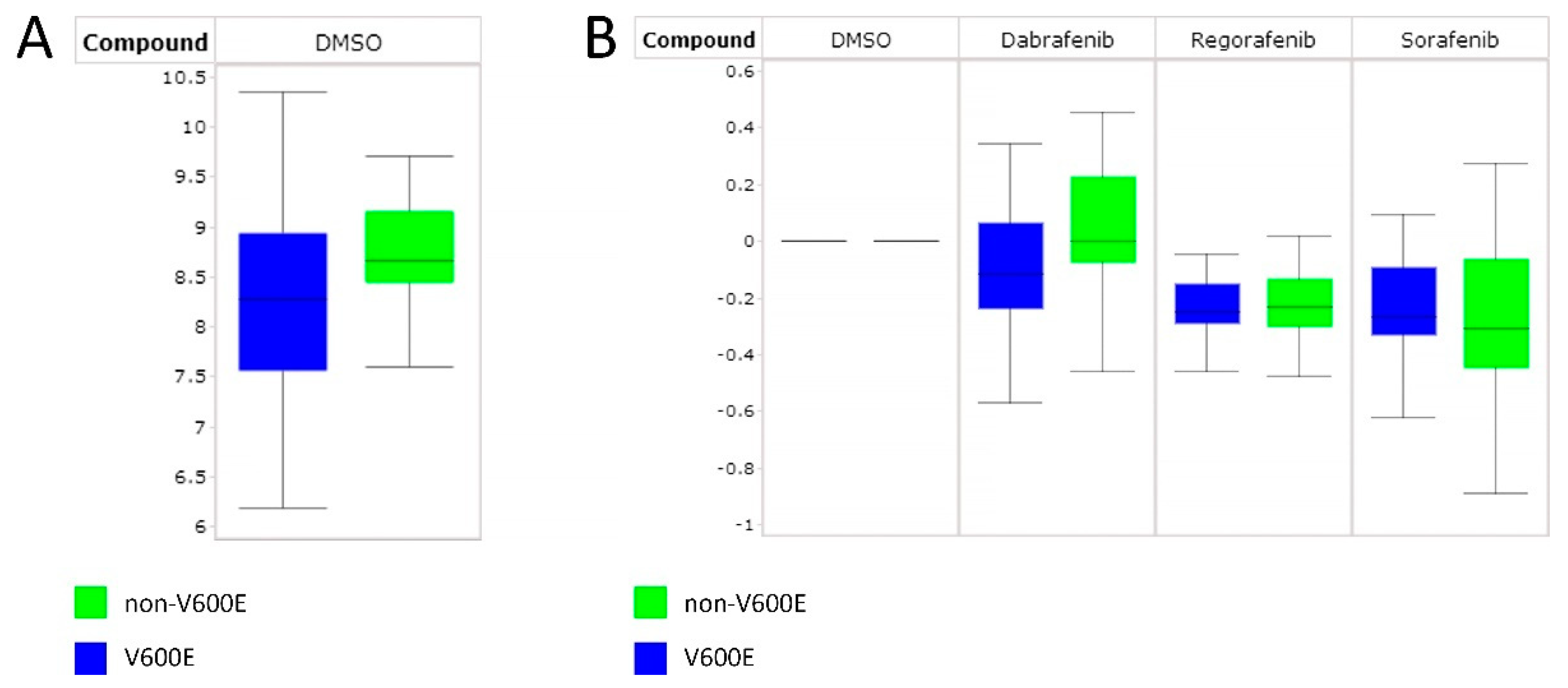
3.4. Differentiating Non-BRAF V600E and BRAF V600E PTC Using RAF Inhibitors
4. Discussion
4.1. Kinase Activity Profiling Correctly Classifies Benign and Malignant Thyroid Tumors
4.2. Molecular Interpretation of Differences between Benign and Malignant Samples
4.3. Distinguishing Non-BRAF V600E from BRAF V600E PTC Using RAF Inhibitors
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bai, Y.; Kakudo, K.; Jung, C.K. Updates in the Pathologic Classification of Thyroid Neoplasms: A Review of the World Health Organization Classification. Endocrinol. Metab. 2020, 35, 696–715. [Google Scholar] [CrossRef] [PubMed]
- Christofer Juhlin, C.; Mete, O.; Baloch, Z.W. The 2022 WHO classification of thyroid tumors: Novel concepts in nomenclature and grading. Endocr. Relat. Cancer 2023, 30, e220293. [Google Scholar] [CrossRef] [PubMed]
- Corver, W.E.; Ruano, D.; Weijers, K.; den Hartog, W.C.; van Nieuwenhuizen, M.P.; de Miranda, N.; van Eijk, R.; Middeldorp, A.; Jordanova, E.S.; Oosting, J.; et al. Genome haploidisation with chromosome 7 retention in oncocytic follicular thyroid carcinoma. PLoS ONE 2012, 7, e38287. [Google Scholar] [CrossRef] [PubMed]
- Corver, W.E.; van Wezel, T.; Molenaar, K.; Schrumpf, M.; van den Akker, B.; van Eijk, R.; Ruano Neto, D.; Oosting, J.; Morreau, H. Near-haploidization significantly associates with oncocytic adrenocortical, thyroid, and parathyroid tumors but not with mitochondrial DNA mutations. Genes Chromosomes Cancer 2014, 53, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Bible, K.C.; Ryder, M. Evolving molecularly targeted therapies for advanced-stage thyroid cancers. Nat. Rev. Clin. Oncol. 2016, 13, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Cabanillas, M.E.; McFadden, D.G.; Durante, C. Thyroid cancer. Lancet 2016, 388, 2783–2795. [Google Scholar] [CrossRef]
- Tesselaar, M.H.; Smit, J.W.; Nagarajah, J.; Netea-Maier, R.T.; Plantinga, T.S. Pathological processes and therapeutic advances in radioiodide refractory thyroid cancer. J. Mol. Endocrinol. 2017, 59, R141–R154. [Google Scholar] [CrossRef]
- Ganly, I.; Makarov, V.; Deraje, S.; Dong, Y.; Reznik, E.; Seshan, V.; Nanjangud, G.; Eng, S.; Bose, P.; Kuo, F.; et al. Integrated Genomic Analysis of Hurthle Cell Cancer Reveals Oncogenic Drivers, RecuR.R.ent Mitochondrial Mutations, and Unique Chromosomal Landscapes. Cancer Cell 2018, 34, 256–270.e255. [Google Scholar] [CrossRef]
- Valero, C.; Ganly, I.; Shah, J.P. Head and neck paragangliomas: 30-year experience. Head Neck 2020, 42, 10. [Google Scholar] [CrossRef]
- Caronia, L.M.; Phay, J.E.; Shah, M.H. Role of BRAF in thyroid oncogenesis. Clin. Cancer Res. 2011, 17, 7511–7517. [Google Scholar] [CrossRef]
- Schneider, T.C.; Abdulrahman, R.M.; Corssmit, E.P.; Morreau, H.; Smit, J.W.; Kapiteijn, E. Long-term analysis of the efficacy and tolerability of sorafenib in advanced radio-iodine refractory differentiated thyroid carcinoma: Final results of a phase II trial. Eur. J. Endocrinol. 2012, 167, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Brose, M.S.; Nutting, C.M.; Jarzab, B.; Elisei, R.; Siena, S.; Bastholt, L.; de la Fouchardiere, C.; Pacini, F.; Paschke, R.; Shong, Y.K.; et al. Sorafenib in radioactive iodine-refractory, locally advanced or metastatic differentiated thyroid cancer: A randomised, double-blind, phase 3 trial. Lancet 2014, 384, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Brose, M.S.; Cabanillas, M.E.; Cohen, E.E.; Wirth, L.J.; Riehl, T.; Yue, H.; Sherman, S.I.; Sherman, E.J. Vemurafenib in patients with BRAF(V600E)-positive metastatic or unresectable papillary thyroid cancer refractory to radioactive iodine: A non-randomised, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016, 17, 1272–1282. [Google Scholar] [CrossRef]
- Huillard, O.; Tenenbaum, F.; Clerc, J.; Goldwasser, F.; Groussin, L. Vemurafenib for BRAFV600E-positive metastatic papillary thyroid cancer. Lancet Oncol. 2016, 17, e468. [Google Scholar] [CrossRef]
- Rothenberg, S.M.; McFadden, D.G.; Palmer, E.L.; Daniels, G.H.; Wirth, L.J. Redifferentiation of iodine-refractory BRAF V600E-mutant metastatic papillary thyroid cancer with dabrafenib. Clin. Cancer Res. 2015, 21, 1028–1035. [Google Scholar] [CrossRef]
- Dunn, L.A.; Sherman, E.J.; Baxi, S.S.; Tchekmedyian, V.; Grewal, R.K.; Larson, S.M.; Pentlow, K.S.; Haque, S.; Tuttle, R.M.; Sabra, M.M.; et al. Vemurafenib Redifferentiation of BRAF Mutant, RAI-Refractory Thyroid Cancers. J. Clin. Endocrinol. Metab. 2019, 104, 1417–1428. [Google Scholar] [CrossRef]
- Falchook, G.S.; Millward, M.; Hong, D.; Naing, A.; Piha-Paul, S.; Waguespack, S.G.; Cabanillas, M.E.; Sherman, S.I.; Ma, B.; Curtis, M.; et al. BRAF inhibitor dabrafenib in patients with metastatic BRAF-mutant thyroid cancer. Thyroid 2015, 25, 71–77. [Google Scholar] [CrossRef]
- Fujiwara, Y.; Yamazaki, N.; Kiyohara, Y.; Yoshikawa, S.; Yamamoto, N.; Tsutsumida, A.; Nokihara, H.; Namikawa, K.; Mukaiyama, A.; Zhang, F.; et al. Safety, tolerability, and pharmacokinetic profile of dabrafenib in Japanese patients with BRAF V600 mutation-positive solid tumors: A phase 1 study. Investig. New Drugs 2017, 36, 10. [Google Scholar] [CrossRef]
- Tchekmedyian, V.; Dunn, L.; Sherman, E.; Baxi, S.S.; Grewal, R.K.; Larson, S.M.; Pentlow, K.S.; Haque, S.; Tuttle, R.M.; Sabra, M.M.; et al. Enhancing Radioiodine Incorporation in BRAF- Mutant, Radioiodine-Refractory Thyroid Cancers with Vemurafenib and the Anti-ErbB3 Monoclonal Antibody CDX-3379: Results of a Pilot Clinical Trial. Thyroid 2022, 32, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Hilhorst, R.; Houkes, L.; Mommersteeg, M.; Musch, J.; van den Berg, A.; Ruijtenbeek, R. Peptide microaR.R.ays for profiling of serine/threonine kinase activity of recombinant kinases and lysates of cells and tissue samples. Methods Mol. Biol. 2013, 977, 259–271. [Google Scholar] [CrossRef]
- van der Tuin, K.; Ventayol Garcia, M.; Corver, W.E.; Khalifa, M.N.; Ruano Neto, D.; Corssmit, E.P.M.; Hes, F.J.; Links, T.P.; Smit, J.W.A.; Plantinga, T.S.; et al. Targetable gene fusions identified in radioactive iodine refractory advanced thyroid carcinoma. Eur. J. Endocrinol. 2019, 180, 235–241. [Google Scholar] [CrossRef] [PubMed]
- van Eijk, R.; Stevens, L.; Morreau, H.; van Wezel, T. Assessment of a fully automated high- throughput DNA extraction method from formalin-fixed, paraffin-embedded tissue for KRAS, and BRAF somatic mutation analysis. Exp. Mol. Pathol. 2013, 94, 121–125. [Google Scholar] [CrossRef]
- Sibinga Mulder, B.G.; Mieog, J.S.D.; Handgraaf, H.J.M.; Farina Sarasqueta, A.; Vasen, H.F.A.; Potjer, T.P.; Swijnenburg, R.-J.; Luelmo, S.A.C.; Feshtali, S.; Inderson, A.; et al. Targeted next- generation sequencing of FNA-derived DNA in pancreatic cancer. J. Clin. Pathol. 2017, 70, 174–178. [Google Scholar] [CrossRef]
- Corver, W.E.; Middeldorp, A.; Ter Haar, N.T.; Jordanova, E.S.; van Puijenbroek, M.; van Eijk, R.; Cornelisse, C.J.; Fleuren, G.J.; Morreau, H.; Oosting, J.; et al. Genome-wide allelic state analysis on flow-sorted tumor fractions provides an accurate measure of chromosomal abeR.R.ations. Cancer Res. 2008, 68, 10333–10340. [Google Scholar] [CrossRef] [PubMed]
- Barker, M.; Rayens, W. Partial least squares for discrimination. J. Chemometr. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Boulesteix, A.L.; Strobl, C.; Augustin, T.; Daumer, M. Evaluating microaR.R.ay-based classifiers: An overview. Cancer Inform. 2008, 6, 77–97. [Google Scholar] [CrossRef] [PubMed]
- Arni, S.; Le, T.H.N.; de Wijn, R.; Garcia-Villegas, R.; Dankers, M.; Weder, W.; Hillinger, S. Ex vivo multiplex profiling of protein tyrosine kinase activities in early stages of human lung adenocarcinoma. Oncotarget 2017, 8, 68599–68613. [Google Scholar] [CrossRef]
- Benjamini, Y.; Drai, D.; Elmer, G.; Kafkafi, N.; Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001, 125, 279–284. [Google Scholar] [CrossRef]
- Chirumamilla, C.S.; Fazil, M.; Perez-Novo, C.; Rangarajan, S.; de Wijn, R.; Ramireddy, P.; Verma, N.K.; Vanden Berghe, W. Profiling Activity of Cellular Kinases in Migrating T-Cells. Methods Mol. Biol. 2019, 1930, 99–113. [Google Scholar] [CrossRef]
- Pearson, W.R. Selecting the Right Similarity-Scoring Matrix. Curr. Protoc. Bioinform. 2013, 43, 9. [Google Scholar] [CrossRef]
- Metz, K.S.; Deoudes, E.M.; Berginski, M.E.; Jimenez-Ruiz, I.; Aksoy, B.A.; Hammerbacher, J.; Gomez, S.M.; Phanstiel, D.H. Coral: Clear and Customizable Visualization of Human Kinome Data. Cell Syst. 2018, 7, 347–350.e341. [Google Scholar] [CrossRef] [PubMed]
- Corver, W.; Demmers, J.; Oosting, J.; Sahraeian, S.; Boot, A.; Ruano, D.; van Wezel, T.; Morreau, H. ROS-induced near-homozygous genomes in thyroid cancer. Endocr. Relat. Cancer 2018, 25, 15. [Google Scholar] [CrossRef] [PubMed]
- de Koster, E.J.; Corver, W.E.; de Geus-Oei, L.F.; Oyen, W.J.G.; Ruano, D.; Schepers, A.; Snel, M.; van Wezel, T.; Vriens, D.; Morreau, J. A clinically applicable molecular classification of Hürthle cell thyroid nodules. Endocr. Relat. Cancer 2023, 30, e230047. [Google Scholar] [CrossRef] [PubMed]
- Krayem, M.; Aftimos, P.; Najem, A.; van den Hooven, T.; van den Berg, A.; Hovestad-Bijl, L.; de Wijn, R.; Hilhorst, R.; Ruijtenbeek, R.; Sabbah, M.; et al. Kinome Profiling to Predict Sensitivity to MAPK Inhibition in Melanoma and to Provide New Insights into Intrinsic and Acquired Mechanism of Resistance. Cancers 2020, 12, 512. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014, 159, 676–690. [Google Scholar] [CrossRef]
- Cancer Tissue Specific Score. Available online: https://www.proteinatlas.org/search/cancercategoryrna%3Athyroid+cancer%3BCancer+enriched%2CGroup+enriched%2CCancer+enhanced+AND+sortby%3Atissue+specific+score/6 (accessed on 19 January 2023).
- PRKCQ. Available online: https://www.proteinatlas.org/ENSG00000065675-PRKCQ (accessed on 19 January 2023).
- Available online: https://www.proteinatlas.org/ENSG00000184304-PRKD1/pathology (accessed on 19 January 2023).
- Saavedra, A.P.; Tsygankova, O.M.; Prendergast, G.V.; Dworet, J.H.; Cheng, G.; Meinkoth, J.L. Role of cAMP, PKA and Rap1A in thyroid follicular cell survival. Oncogene 2002, 21, 778–788. [Google Scholar] [CrossRef] [PubMed]
- Nikiforova, M.N.; Wald, A.I.; Roy, S.; Durso, M.B.; Nikiforov, Y.E. Targeted next-generation sequencing panel (ThyroSeq) for detection of mutations in thyroid cancer. J. Clin. Endocrinol. Metab. 2013, 98, E1852–E1860. [Google Scholar] [CrossRef]
- Salabe, G.B. Pathogenesis of thyroid nodules: Histological classification? Biomed. Pharmacother. 2001, 55, 39–53. [Google Scholar] [CrossRef]
- Del Gobbo, A.; Peverelli, E.; Treppiedi, D.; Lania, A.; Mantovani, G.; Ferrero, S. Expression of protein kinase A regulatory subunits in benign and malignant human thyroid tissues: A systematic review. Exp. Cell Res. 2016, 346, 85–90. [Google Scholar] [CrossRef]
- Kari, S.; Vasko, V.V.; Priya, S.; Kirschner, L.S. PKA Activates AMPK Through LKB1 Signaling in Follicular Thyroid Cancer. Front. Endocrinol. 2019, 10, 769. [Google Scholar] [CrossRef]
- Miasaki, F.Y.; Fuziwara, C.S.; Carvalho, G.A.; Kimura, E.T. Genetic Mutations and Variants in the Susceptibility of Familial Non-Medullary Thyroid Cancer. Genes 2020, 11, 1364. [Google Scholar] [CrossRef] [PubMed]
- Huo, K.G.; Notsuda, H.; Fang, Z.; Liu, N.F.; Gebregiworgis, T.; Li, Q.; Pham, N.A.; Li, M.; Liu, N.; Shepherd, F.A.; et al. Lung Cancer Driven by BRAF(G469V) Mutation Is Targetable by EGFR Kinase Inhibitors. J. Thorac. Oncol. 2022, 17, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Piperno-Neumann, S.; Carlino, M.S.; Boni, V.; Loirat, D.; Speetjens, F.M.; Park, J.J.; Calvo, E.; Carvajal, R.D.; Nyakas, M.; Gonzalez-Maffe, J.; et al. A phase I trial of LXS196, a protein kinase C (PKC) inhibitor, for metastatic uveal melanoma. Br. J. Cancer 2023, 128, 1040–1051. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.; Chen, H.; Feng, Y.; Cui, B.; Kang, Y.; Zhang, P.; Luo, M.; Chen, J. Small-Molecule Inhibitor Targeting Protein Kinase D: A Potential Therapeutic Strategy. Front. Oncol. 2021, 11, 680221. [Google Scholar] [CrossRef]
- Long, G.V.; Stroyakovskiy, D.; Gogas, H.; Levchenko, E.; de Braud, F.; Larkin, J.; Garbe, C.; Jouary, T.; Hauschild, A.; Grob, J.J.; et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N. Engl. J. Med. 2014, 371, 1877–1888. [Google Scholar] [CrossRef]
- Kopetz, S.; Desai, J.; Chan, E.; Hecht, J.R.; O’Dwyer, P.J.; Maru, D.; Morris, V.; Janku, F.; Dasari, A.; Chung, W.; et al. Phase II Pilot Study of Vemurafenib in Patients with Metastatic BRAF-Mutated Colorectal Cancer. J. Clin. Oncol. 2015, 33, 4032–4038. [Google Scholar] [CrossRef]
- Corcoran, R.B.; Atreya, C.E.; Falchook, G.S.; Kwak, E.L.; Ryan, D.P.; Bendell, J.C.; Hamid, O.; Messersmith, W.A.; Daud, A.; Kurzrock, R.; et al. Combined BRAF and MEK Inhibition with Dabrafenib and Trametinib in BRAF V600-Mutant Colorectal Cancer. J. Clin. Oncol. 2015, 33, 4023–4031. [Google Scholar] [CrossRef]
- Subbiah, V.; Kreitman, R.J.; Wainberg, Z.A.; Cho, J.Y.; Schellens, J.H.M.; Soria, J.C.; Wen, P.Y.; Zielinski, C.; Cabanillas, M.E.; Urbanowitz, G.; et al. Dabrafenib and Trametinib Treatment in Patients with Locally Advanced or Metastatic BRAF V600-Mutant Anaplastic Thyroid Cancer. J. Clin. Oncol. 2018, 36, 7–13. [Google Scholar] [CrossRef]
- Iravani, A.; Solomon, B.; Pattison, D.A.; Jackson, P.; Ravi Kumar, A.; Kong, G.; Hofman, M.S.; Akhurst, T.; Hicks, R.J. Mitogen-Activated Protein Kinase Pathway Inhibition for Redifferentiation of Radioiodine Refractory Differentiated Thyroid Cancer: An Evolving Protocol. Thyroid 2019, 29, 1634–1645. [Google Scholar] [CrossRef]
- Jafri, S.; Yaqub, A. Redifferentiation of BRAF V600E-Mutated Radioiodine Refractory Metastatic Papillary Thyroid Cancer After Treatment with Dabrafenib and Trametinib. Cureus 2021, 13, e17488. [Google Scholar] [CrossRef]
- Ullmann, T.M.; Liang, H.; Moore, M.D.; Al-Jamed, I.; Gray, K.D.; Limberg, J.; Stefanova, D.; Buicko, J.L.; Finnerty, B.; Beninato, T.; et al. Dual inhibition of BRAF and MEK increases expression of sodium iodide symporter in patient-derived papillary thyroid cancer cells in vitro. Surgery 2020, 167, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Gopal, R.K.; Kubler, K.; Calvo, S.E.; Polak, P.; Livitz, D.; Rosebrock, D.; Sadow, P.M.; Campbell, B.; Donovan, S.E.; Amin, S.; et al. Widespread Chromosomal Losses and Mitochondrial DNA Alterations as Genetic Drivers in Hurthle Cell Carcinoma. Cancer Cell 2018, 34, 242–255.e245. [Google Scholar] [CrossRef] [PubMed]


| Thyroid Presence | ||||||||
|---|---|---|---|---|---|---|---|---|
| Rank | Kinase | Gene Name | UniProt ID | Kinase Score | Normal Tissue | Cancer | Reliability Score * | Remarks |
| 1 | RSK2 | RPS6KA3 | P51812 | 5.09 | yes | yes | approved | |
| 2 | MSK1 | RPS6KA5 | O75582 | 4.74 | yes | yes | approved | |
| 3 | PKC[epsilon] | PRKCE | Q02156 | 4.46 | yes | yes | enhanced | |
| 4 | PKA[alpha] | PRKACA | P17612 | 4.66 | yes | yes | approved | |
| 5 | Pim1 | PIM1 | P11309 | 4.38 | low | low | uncertain | |
| 6 | SGK2 | SGK2 | Q9HBY8 | 3.82 | low | low | uncertain | |
| 7 | ANP[alpha] | NPR1 | P16066 | 3.98 | yes | yes | na | Has kinase-like homology domain |
| 8 | p70S6K[beta] | RPS6KB2 | Q9UBS0 | 3.90 | yes | yes | approved | |
| 9 | PKC[gamma] | PRKCG | P05129 | 3.89 | low | low | enhanced | |
| 10 | Pim3 | PIM3 | Q86V86 | 4.06 | yes | yes | na | |
| 11 | PRKX | PRKX | P51817 | 3.96 | yes | yes | supported | Thyroid cancer enhanced |
| 12 | AurA/Aur2 | AURKA | O14965 | 3.77 | low | low | enhanced | |
| 13 | Pim2 | PIM2 | Q9P1W9 | 3.71 | low | low | na | |
| 14 | MSK2 | RPS6KA4/RSKB | O75676 | 3.70 | yes | yes | approved | |
| 15 | CHK2 | CHEK2 | O96017 | 3.64 | yes | yes | enhanced | |
| 16 | GSK3[beta] | GSK3B | P49841 | 3.66 | yes | yes | approved | |
| 17 | PKC[theta] | PRKCQ | Q04759 | 3.62 | yes | yes | na | Unfavorable prognostic marker |
| 18 | PKD1 | PRKD1 | Q15139 | 3.62 | yes | yes | uncertain | Unfavorable prognostic marker |
| 19 | PKC[delta] | PRKCD | Q05655 | 3.74 | yes | yes | enhanced | |
| 20 | Akt1/PKB[alpha] | PKB (alpha) | P31749 | 3.61 | yes | yes | supported | |
| 21 | Akt2/PKB[beta] | PKB (beta) | P31751 | 3.58 | yes | yes | uncertain | |
| 22 | CDKL5 | CDKL5 | O76039 | 3.59 | yes | yes | approved | |
| 23 | GSK3[alpha] | GSK3A | P49840 | 3.45 | yes | yes | approved | |
| 24 | AurB/Aur1 | AURKB | Q96GD4 | 3.49 | low | low | enhanced | |
| 25 | p70S6K | RPS6KB1 | P23443 | 3.45 | yes | yes | approved | |
| 26 | RSKL2 | RPS6KL1 | Q9Y6S9 | 3.49 | low | low | approved | |
| 27 | TBK1 | TBK1 | Q9UHD2 | 3.36 | yes | yes | uncertain | |
| 28 | PKC[alpha] | PRKCA | P17252 | 3.47 | low | low | enhanced | |
| 29 | PKC[beta] | PRKCB | P05771 | 3.46 | yes | yes | approved | |
| 30 | CaMK4 | CAMK4 | Q16566 | 3.49 | low | low | approved | |
| 31 | PRKY | PRKY | O43930 | 3.28 | na | na | na | |
| 32 | PKC[zeta] | PRKCZ | Q05513 | 3.40 | yes | yes | enhanced | |
| 33 | ADCK3 | COQ8A | Q8NI60 | 3.27 | na | na | na | |
| 34 | PKG1 | PRKG1 | Q13976 | 3.48 | yes | yes | enhanced | |
| 35 | PKN1/PRK1 | PKN1 | Q16512 | 3.38 | yes | yes | approved | |
| 36 | PKC[eta] | PRKCH | P24723 | 3.30 | yes | yes | approved | |
| 37 | mTOR/FRAP | FRAP | P42345 | 3.31 | yes | yes | approved | |
| 38 | RSK3 | RPS6KA1 | Q15418 | 3.30 | yes | yes | approved | |
| 39 | RSK1/p90RSK | RPS6KA2 | Q15349 | 3.27 | yes | yes | enhanced | Thyroid cancer enhanced |
| 40 | AlphaK1 | ALPK3 | Q96L96 | 3.25 | low | yes | uncertain |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hilhorst, R.; van den Berg, A.; Boender, P.; van Wezel, T.; Kievits, T.; de Wijn, R.; Ruijtenbeek, R.; Corver, W.E.; Morreau, H. Differentiating Benign from Malignant Thyroid Tumors by Kinase Activity Profiling and Dabrafenib BRAF V600E Targeting. Cancers 2023, 15, 4477. https://doi.org/10.3390/cancers15184477
Hilhorst R, van den Berg A, Boender P, van Wezel T, Kievits T, de Wijn R, Ruijtenbeek R, Corver WE, Morreau H. Differentiating Benign from Malignant Thyroid Tumors by Kinase Activity Profiling and Dabrafenib BRAF V600E Targeting. Cancers. 2023; 15(18):4477. https://doi.org/10.3390/cancers15184477
Chicago/Turabian StyleHilhorst, Riet, Adrienne van den Berg, Piet Boender, Tom van Wezel, Tim Kievits, Rik de Wijn, Rob Ruijtenbeek, Willem E. Corver, and Hans Morreau. 2023. "Differentiating Benign from Malignant Thyroid Tumors by Kinase Activity Profiling and Dabrafenib BRAF V600E Targeting" Cancers 15, no. 18: 4477. https://doi.org/10.3390/cancers15184477
APA StyleHilhorst, R., van den Berg, A., Boender, P., van Wezel, T., Kievits, T., de Wijn, R., Ruijtenbeek, R., Corver, W. E., & Morreau, H. (2023). Differentiating Benign from Malignant Thyroid Tumors by Kinase Activity Profiling and Dabrafenib BRAF V600E Targeting. Cancers, 15(18), 4477. https://doi.org/10.3390/cancers15184477







