Three-Dimensional Spheroid Configurations and Cellular Metabolic Properties of Oral Squamous Carcinomas Are Possible Pharmacological and Pathological Indicators
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Establishment of Oral Squamous Carcinoma (OSCC) or Oral Adenosquamous Carcinoma (OASC) Cell Lines and Their 2D and 3D Cell Cultures
2.2. Real-Time Analysis of the Cellular Metabolic Functions of Various OSCC and OASC Cell Lines
2.3. Morphological Analyses of the 3D Spheroids Derived from Various OSCC and OASC Cells
2.4. Assays for Cisplatin-Induced Cytotoxicity
2.5. Assays for Trypsin-Induced Dispersion
2.6. Other Methods
3. Results
3.1. Cellular Metabolic Aspects of 2D Cultured Four Established OSCC or OASC Cell Lines
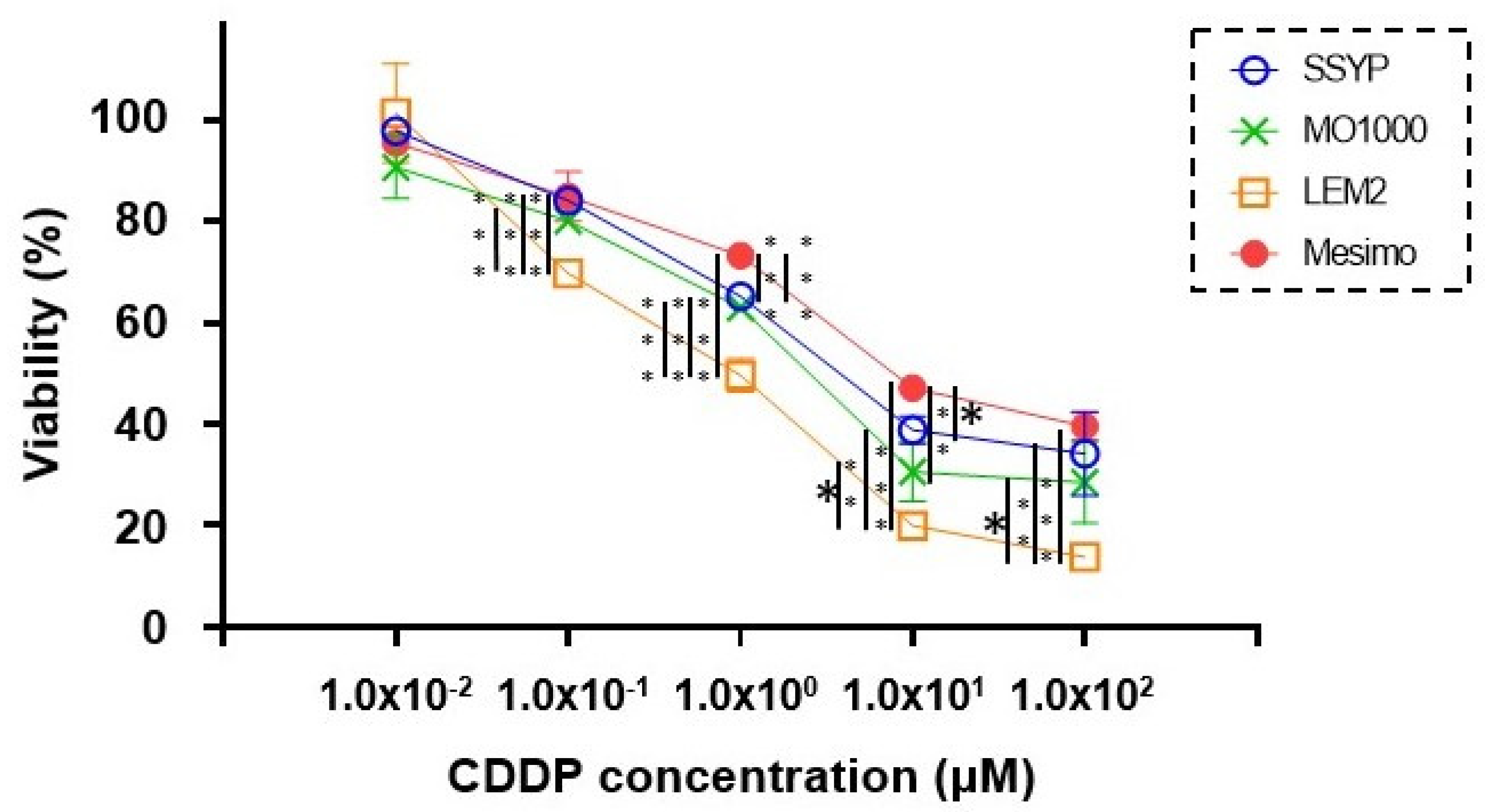
3.2. Cytotoxicity of 2D Cultured Four Established OSCC or OASC Cell Lines against Cisplatin (CDDP)
3.3. D Spheroid Configurations of Four Established OSCC or OASC Cell Lines
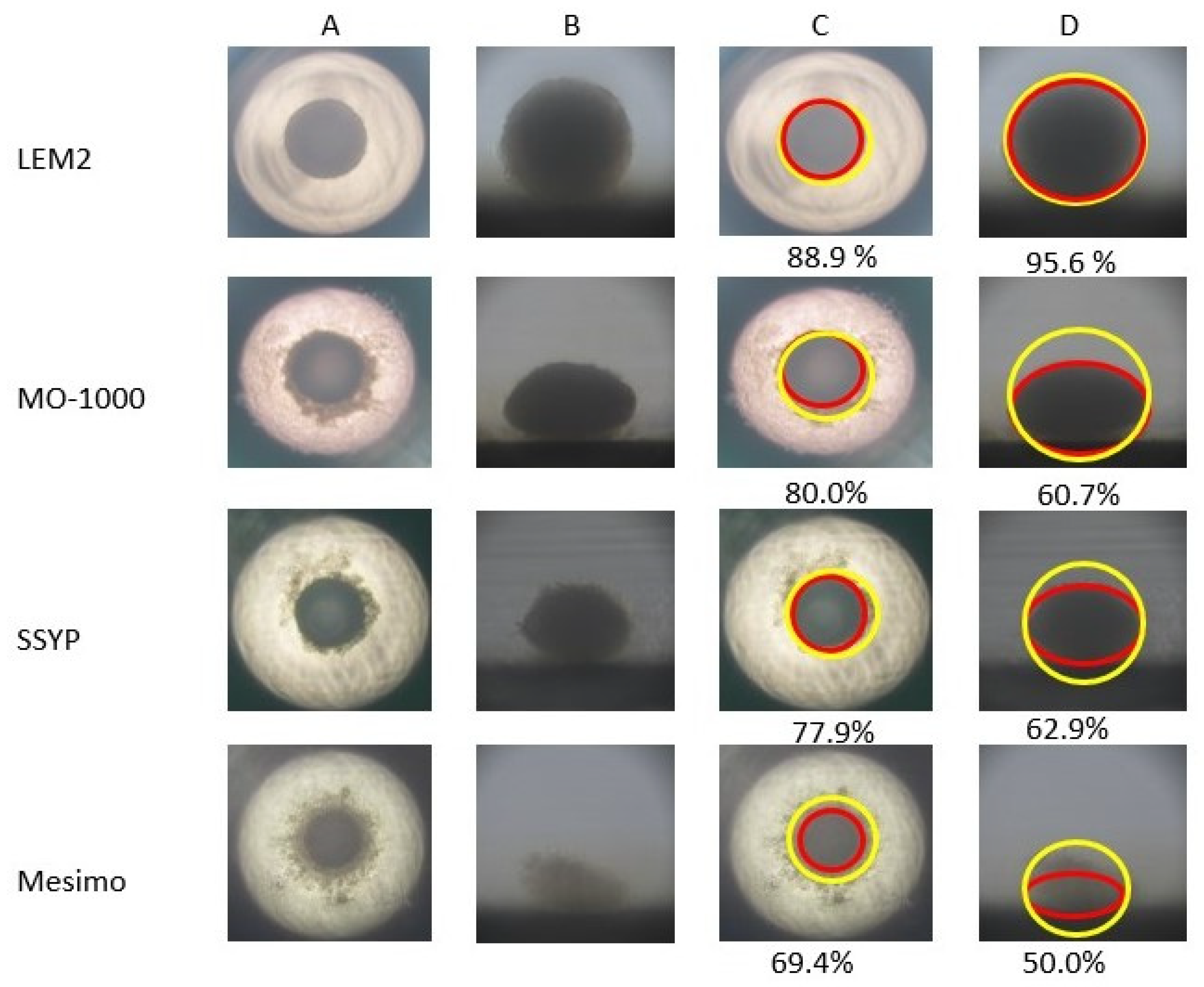
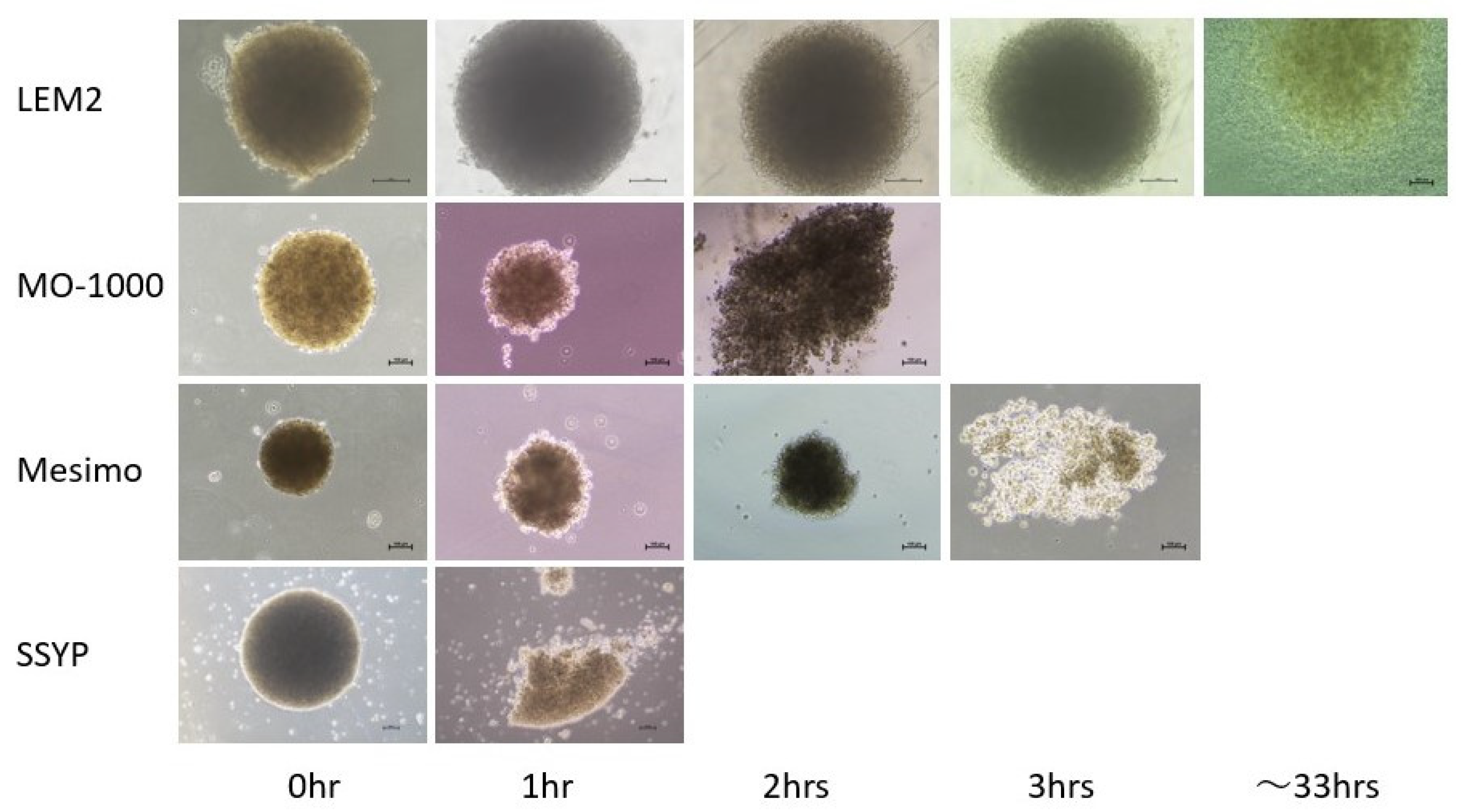
3.4. Trypsin-Induced Dispersion of 2D and 3D Cultures of Four Established OSCC or OASC Cell Lines
3.5. The mRNA Expression of Possibly Related Molecules within the Biological Diversities among 2D and 3D Cultures of the Four Established OSCC or OASC Cell Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- McCullough, M.J.; Prasad, G.; Farah, C.S. Oral mucosal malignancy and potentially malignant lesions: An update on the epidemiology, risk factors, diagnosis and management. Aust. Dent. J. 2010, 55 (Suppl. 1), 61–65. [Google Scholar] [CrossRef]
- Farah, C.S.; Woo, S.B.; Zain, R.B.; Sklavounou, A.; McCullough, M.J.; Lingen, M. Oral cancer and oral potentially malignant disorders. Int. J. Dent. 2014, 2014, 853479. [Google Scholar] [CrossRef]
- Abrahão, R.; Anantharaman, D.; Gaborieau, V.; Abedi-Ardekani, B.; Lagiou, P.; Lagiou, A.; Ahrens, W.; Holcatova, I.; Betka, J.; Merletti, F.; et al. The influence of smoking, age and stage at diagnosis on the survival after larynx, hypopharynx and oral cavity cancers in Europe: The ARCAGE study. Int. J. Cancer 2018, 143, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Irani, S.; Barati, I.; Badiei, M. Periodontitis and oral cancer—Current concepts of the etiopathogenesis. Oncol. Rev. 2020, 14, 465. [Google Scholar] [CrossRef] [PubMed]
- Isola, G.; Polizzi, A.; Patini, R.; Ferlito, S.; Alibrandi, A.; Palazzo, G. Association among serum and salivary A. actinomycetemcomitans specific immunoglobulin antibodies and periodontitis. BMC Oral Health 2020, 20, 283. [Google Scholar] [CrossRef]
- Isola, G.; Lo Giudice, A.; Polizzi, A.; Alibrandi, A.; Murabito, P.; Indelicato, F. Identification of the different salivary Interleukin-6 profiles in patients with periodontitis: A cross-sectional study. Arch. Oral Biol. 2021, 122, 104997. [Google Scholar] [CrossRef]
- Pulte, D.; Brenner, H. Changes in survival in head and neck cancers in the late 20th and early 21st century: A period analysis. Oncologist 2010, 15, 994–1001. [Google Scholar] [CrossRef]
- Bissell, M.J.; Radisky, D. Putting tumours in context. Nat. Rev. Cancer 2001, 1, 46–54. [Google Scholar] [CrossRef]
- Horvath, P.; Aulner, N.; Bickle, M.; Davies, A.M.; Nery, E.D.; Ebner, D.; Montoya, M.C.; Östling, P.; Pietiäinen, V.; Price, L.S.; et al. Screening out irrelevant cell-based models of disease. Nat. Rev. Drug Discov. 2016, 15, 751–769. [Google Scholar] [CrossRef] [PubMed]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Miki, Y.; Ono, K.; Hata, S.; Suzuki, T.; Kumamoto, H.; Sasano, H. The advantages of co-culture over mono cell culture in simulating in vivo environment. J. Steroid Biochem. Mol. Biol. 2012, 131, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Shang, M.; Soon, R.H.; Lim, C.T.; Khoo, B.L.; Han, J. Microfluidic modelling of the tumor microenvironment for anti-cancer drug development. Lab Chip 2019, 19, 369–386. [Google Scholar] [CrossRef]
- Yamada, K.M.; Cukierman, E. Modeling tissue morphogenesis and cancer in 3D. Cell 2007, 130, 601–610. [Google Scholar] [CrossRef]
- Zanoni, M.; Pignatta, S.; Arienti, C.; Bonafè, M.; Tesei, A. Anticancer drug discovery using multicellular tumor spheroid models. Expert Opin. Drug Discov. 2019, 14, 289–301. [Google Scholar] [CrossRef]
- Santini, M.T.; Rainaldi, G. Three-dimensional spheroid model in tumor biology. Pathobiol. J. Immunopathol. Mol. Cell. Biol. 1999, 67, 148–157. [Google Scholar] [CrossRef]
- Katt, M.E.; Placone, A.L.; Wong, A.D.; Xu, Z.S.; Searson, P.C. In Vitro Tumor Models: Advantages, Disadvantages, Variables, and Selecting the Right Platform. Front. Bioeng. Biotechnol. 2016, 4, 12. [Google Scholar] [CrossRef]
- Hikage, F.; Atkins, S.; Kahana, A.; Smith, T.J.; Chun, T.H. HIF2A-LOX Pathway Promotes Fibrotic Tissue Remodeling in Thyroid-Associated Orbitopathy. Endocrinology 2019, 160, 20–35. [Google Scholar] [CrossRef]
- Watanabe, M.; Ida, Y.; Furuhashi, M.; Tsugeno, Y.; Ohguro, H.; Hikage, F. Screening of the Drug-Induced Effects of Prostaglandin EP2 and FP Agonists on 3D Cultures of Dexamethasone-Treated Human Trabecular Meshwork Cells. Biomedicines 2021, 9, 930. [Google Scholar] [CrossRef]
- Oouchi, Y.; Watanabe, M.; Ida, Y.; Ohguro, H.; Hikage, F. Rosiglitasone and ROCK Inhibitors Modulate Fibrogenetic Changes in TGF-β2 Treated Human Conjunctival Fibroblasts (HconF) in Different Manners. Int. J. Mol. Sci. 2021, 22, 7335. [Google Scholar] [CrossRef]
- Ichioka, H.; Hirohashi, Y.; Sato, T.; Furuhashi, M.; Watanabe, M.; Ida, Y.; Hikage, F.; Torigoe, T.; Ohguro, H. G-Protein-Coupled Receptors Mediate Modulations of Cell Viability and Drug Sensitivity by Aberrantly Expressed Recoverin 3 within A549 Cells. Int. J. Mol. Sci. 2023, 24, 771. [Google Scholar] [CrossRef]
- Ohguro, H.; Watanabe, M.; Sato, T.; Hikage, F.; Furuhashi, M.; Okura, M.; Hida, T.; Uhara, H. 3D Spheroid Configurations Are Possible Indictors for Evaluating the Pathophysiology of Melanoma Cell Lines. Cells 2023, 12, 759. [Google Scholar] [CrossRef] [PubMed]
- Ida, Y.; Hikage, F.; Itoh, K.; Ida, H.; Ohguro, H. Prostaglandin F2α agonist-induced suppression of 3T3-L1 cell adipogenesis affects spatial formation of extra-cellular matrix. Sci. Rep. 2020, 10, 7958. [Google Scholar] [CrossRef]
- Marconi, A.; Quadri, M.; Saltari, A.; Pincelli, C. Progress in melanoma modelling in vitro. Exp. Dermatol. 2018, 27, 578–586. [Google Scholar] [CrossRef]
- Yamamoto, E.; Miyakawa, A.; Kohama, G. Mode of invasion and lymph node metastasis in squamous cell carcinoma of the oral cavity. Head Neck Surg. 1984, 6, 938–947. [Google Scholar] [CrossRef]
- Ohguro, H.; Ida, Y.; Hikage, F.; Umetsu, A.; Ichioka, H.; Watanabe, M.; Furuhashi, M. STAT3 Is the Master Regulator for the Forming of 3D Spheroids of 3T3-L1 Preadipocytes. Cells 2022, 11, 300. [Google Scholar] [CrossRef] [PubMed]
- Akama, T.; Leung, B.M.; Labuz, J.; Takayama, S.; Chun, T.H. Designing 3-D Adipospheres for Quantitative Metabolic Study. Methods Mol. Biol. 2017, 1566, 177–183. [Google Scholar] [PubMed]
- Ito, T.; Ohguro, H.; Mamiya, K.; Ohguro, I.; Nakazawa, M. Effects of antiglaucoma drops on MMP and TIMP balance in conjunctival and subconjunctival tissue. Investig. Ophthalmol. Vis. Sci. 2006, 47, 823–830. [Google Scholar] [CrossRef]
- Datta, P.; Dey, M.; Ataie, Z.; Unutmaz, D.; Ozbolat, I.T. 3D bioprinting for reconstituting the cancer microenvironment. NPJ Precis. Oncol. 2020, 4, 18. [Google Scholar] [CrossRef]
- Tanner, K.; Gottesman, M.M. Beyond 3D culture models of cancer. Sci. Transl. Med. 2015, 7, 283ps9. [Google Scholar] [CrossRef] [PubMed]
- Chitturi Suryaprakash, R.T.; Kujan, O.; Shearston, K.; Farah, C.S. Three-Dimensional Cell Culture Models to Investigate Oral Carcinogenesis: A Scoping Review. Int. J. Mol. Sci. 2020, 21, 9520. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Jiang, J.; Zhou, L.; Huang, Z.; Nice, E.C.; Huang, C.; Fu, L. Mitochondrial adaptation in cancer drug resistance: Prevalence, mechanisms, and management. J. Hematol. Oncol. 2022, 15, 97. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, J.; Chen, H.; Wang, R.; Li, P.; Miao, Y.; Liu, P. Metformin suppresses proliferation and invasion of drug-resistant breast cancer cells by activation of the Hippo pathway. J. Cell. Mol. Med. 2020, 24, 5786–5796. [Google Scholar] [CrossRef] [PubMed]
- Tossetta, G. Metformin Improves Ovarian Cancer Sensitivity to Paclitaxel and Platinum-Based Drugs: A Review of In Vitro Findings. Int. J. Mol. Sci. 2022, 23, 12893. [Google Scholar] [CrossRef]
- Naba, A.; Clauser, K.R.; Ding, H.; Whittaker, C.A.; Carr, S.A.; Hynes, R.O. The extracellular matrix: Tools and insights for the “omics” era. Matrix Biol. 2016, 49, 10–24. [Google Scholar] [CrossRef]
- Theocharis, A.D.; Skandalis, S.S.; Gialeli, C.; Karamanos, N.K. Extracellular matrix structure. Adv. Drug Deliv. Rev. 2016, 97, 4–27. [Google Scholar] [CrossRef]
- Oxburgh, L. The Extracellular Matrix Environment of Clear Cell Renal Cell Carcinoma. Cancers 2022, 14, 4072. [Google Scholar] [CrossRef]
- Avery, D.; Govindaraju, P.; Jacob, M.; Todd, L.; Monslow, J.; Puré, E. Extracellular matrix directs phenotypic heterogeneity of activated fibroblasts. Matrix Biol. 2018, 67, 90–106. [Google Scholar] [CrossRef]
- Lenselink, E.A. Role of fibronectin in normal wound healing. Int. Wound J. 2015, 12, 313–316. [Google Scholar] [CrossRef]
- George, E.L.; Georges-Labouesse, E.N.; Patel-King, R.S.; Rayburn, H.; Hynes, R.O. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development 1993, 119, 1079–1091. [Google Scholar] [CrossRef]
- Pankov, R.; Yamada, K.M. Fibronectin at a glance. J. Cell Sci. 2002, 115 Pt 20, 3861–3863. [Google Scholar] [CrossRef]
- To, W.S.; Midwood, K.S. Plasma and cellular fibronectin: Distinct and independent functions during tissue repair. Fibrogenesis Tissue Repair 2011, 4, 21. [Google Scholar] [CrossRef] [PubMed]
- Midwood, K.S.; Williams, L.V.; Schwarzbauer, J.E. Tissue repair and the dynamics of the extracellular matrix. Int. J. Biochem. Cell Biol. 2004, 36, 1031–1037. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, S.; Ross, K.N.; Lander, E.S.; Golub, T.R. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 2003, 33, 49–54. [Google Scholar] [CrossRef]
- Ramakrishnan, V.; Bhaskar, V.; Law, D.A.; Wong, M.H.; DuBridge, R.B.; Breinberg, D.; O’Hara, C.; Powers, D.B.; Liu, G.; Grove, J.; et al. Preclinical evaluation of an anti-alpha5beta1 integrin antibody as a novel anti-angiogenic agent. J. Exp. Ther. Oncol. 2006, 5, 273–286. [Google Scholar] [PubMed]
- Topalovski, M.; Brekken, R.A. Matrix control of pancreatic cancer: New insights into fibronectin signaling. Cancer Lett. 2016, 381, 252–258. [Google Scholar] [CrossRef]
- Murakami, M.; Mimaki, S.; Saitou, Y.; Wada, Y.; Nishiyama, Y.; Yasuda, N.; Murakami, Y. Immunohistological investigation of type IV collagen in the basement membrane surrounding the cancer nest (cancer nest membrane) of head and neck squamous cell carcinoma--its relation to frequency of cervical lymph node metastasis. Nihon Jibiinkoka Gakkai Kaiho 1992, 95, 1773–1784. [Google Scholar] [CrossRef] [PubMed]
- Cescon, M.; Gattazzo, F.; Chen, P.; Bonaldo, P. Collagen VI at a glance. J. Cell Sci. 2015, 128, 3525–3531. [Google Scholar] [CrossRef]
- Bernardi, P.; Bonaldo, P. Dysfunction of mitochondria and sarcoplasmic reticulum in the pathogenesis of collagen VI muscular dystrophies. Ann. N. Y. Acad. Sci. 2008, 1147, 303–311. [Google Scholar] [CrossRef]
- Chen, P.; Cescon, M.; Bonaldo, P. Collagen VI in cancer and its biological mechanisms. Trends Mol. Med. 2013, 19, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Willumsen, N.; Bager, C.; Karsdal, M.A. Matrix Metalloprotease Generated Fragments of Type VI Collagen Have Serum Biomarker Potential in Cancer—A Proof of Concept Study. Transl. Oncol. 2019, 12, 693–698. [Google Scholar] [CrossRef] [PubMed]


| Cell Line | Tumor Site | Histological Type | TNM | Stage | Pathological Grade | YK Classification of Invasion |
|---|---|---|---|---|---|---|
| SSYP | Oral floor | OSCC | T4aN2cM0 | IVA | 1 (well differentiated) | 3 |
| MO-1000 | Gingiva | OSCC | T4aN2cM0 | IVA | 2 (moderately differentiated) | 3 |
| LEM | Gingiva | OSCC | T2N0M0 | II | 2 (moderately differentiated) | 4C |
| Mesimo | Palate | OAdSCC | T3N3bM0 | IVB | 3 (poorly differentiated) | 4D |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miyamoto, S.; Nishikiori, N.; Sato, T.; Watanabe, M.; Umetsu, A.; Tsugeno, Y.; Hikage, F.; Sasaya, T.; Kato, H.; Ogi, K.; et al. Three-Dimensional Spheroid Configurations and Cellular Metabolic Properties of Oral Squamous Carcinomas Are Possible Pharmacological and Pathological Indicators. Cancers 2023, 15, 2793. https://doi.org/10.3390/cancers15102793
Miyamoto S, Nishikiori N, Sato T, Watanabe M, Umetsu A, Tsugeno Y, Hikage F, Sasaya T, Kato H, Ogi K, et al. Three-Dimensional Spheroid Configurations and Cellular Metabolic Properties of Oral Squamous Carcinomas Are Possible Pharmacological and Pathological Indicators. Cancers. 2023; 15(10):2793. https://doi.org/10.3390/cancers15102793
Chicago/Turabian StyleMiyamoto, Sho, Nami Nishikiori, Tatsuya Sato, Megumi Watanabe, Araya Umetsu, Yuri Tsugeno, Fumihito Hikage, Takashi Sasaya, Hirotaka Kato, Kazuhiro Ogi, and et al. 2023. "Three-Dimensional Spheroid Configurations and Cellular Metabolic Properties of Oral Squamous Carcinomas Are Possible Pharmacological and Pathological Indicators" Cancers 15, no. 10: 2793. https://doi.org/10.3390/cancers15102793






