Tumor Treating Fields (TTFields) Concomitant with Sorafenib Inhibit Hepatocellular Carcinoma In Vitro and In Vivo
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Cultures
2.3. In Vitro Experiments
2.4. Flow Cytometry Analysis of Cell Count, Granularity and Apoptosis
2.5. Colony Forming Assay and Overall Effect Calculation
2.6. LC3 Foci Detection by Fluorescent Microscopy, In Vitro
2.7. Western Blot Analysis
2.8. In Vivo Experiment
2.9. MRI Tumor Volume Assessment
2.10. Immunohistochemistry Analysis
2.11. LC3 Foci Detection by Fluorescent Microscopy, In Vivo
2.12. Statistical Analysis
3. Results
3.1. Efficacy of TTFields in HCC Cells
3.2. TTFields Elevate Autophagy in HCC Cell Lines
3.3. Combination of TTFields with Sorafenib Enhances Treatment Efficacy In Vitro
3.4. Autophagy–Apoptosis Interplay For Treatment with Concomitant TTFields and Sorafenib
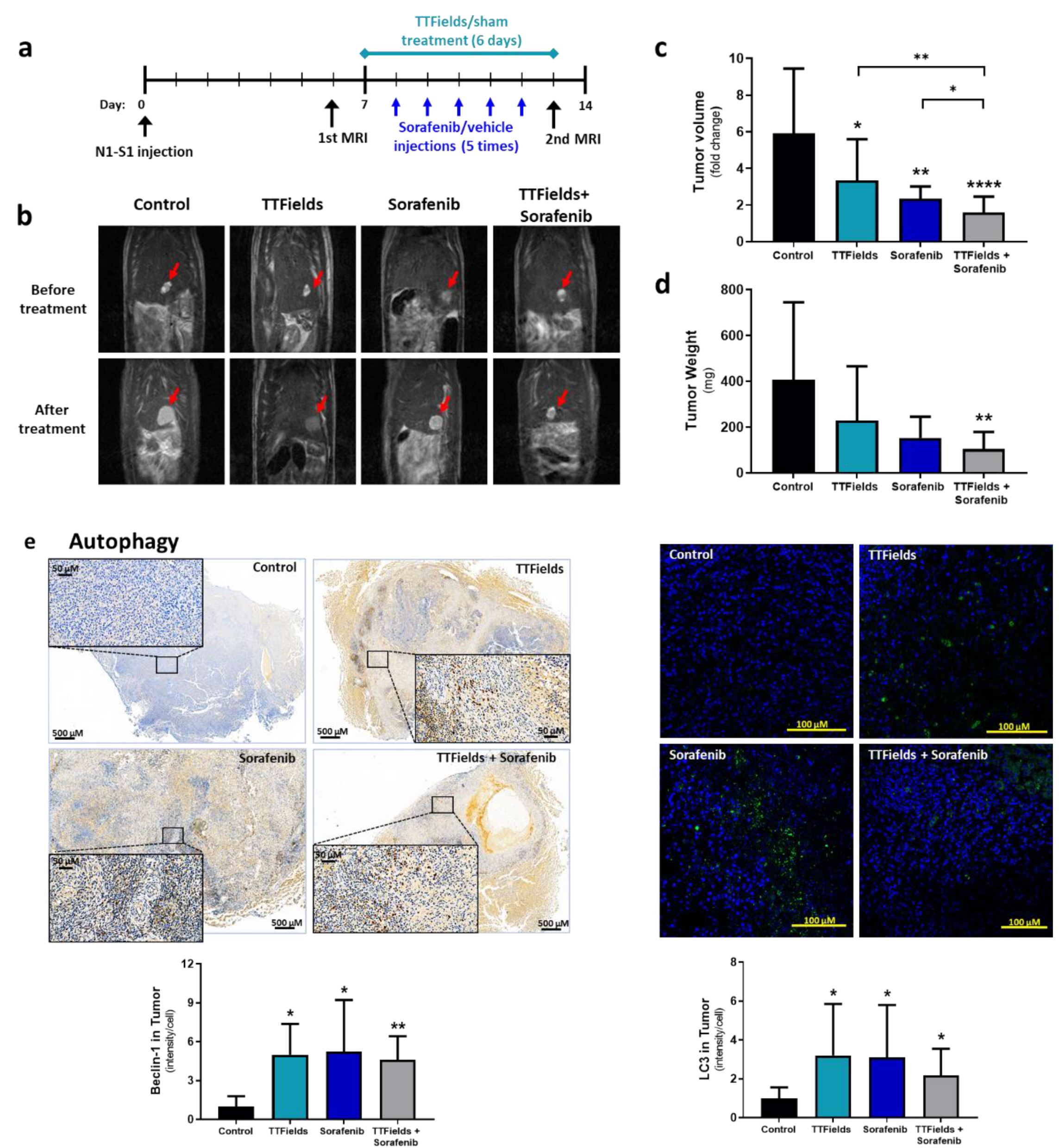
3.5. Concomitant TTFields with Sorafenib Enhances Treatment Efficacy In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liao, J.-Z.; He, X.-X.; Li, P.-Y. The role of autophagy in hepatocellular carcinoma: Friend or foe. Oncotarget 2017, 8, 57707–57722. [Google Scholar] [CrossRef] [PubMed]
- Deng, G.-L.; Zeng, S.; Shen, H. Chemotherapy and target therapy for hepatocellular carcinoma: New advances and challenges. World J. Hepatol. 2015, 7, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Cao, Y.; Chen, C.; Zhang, X.; McNabola, A.; Wilkie, D.; Wilhelm, S.; Lynch, M.; Carter, C. Sorafenib Blocks the RAF/MEK/ERK Pathway, Inhibits Tumor Angiogenesis, and Induces Tumor Cell Apoptosis in Hepatocellular Carcinoma Model PLC/PRF/5. Cancer Res. 2006, 66, 11851–11858. [Google Scholar] [CrossRef]
- Ghavimi, S.; Apfel, T.; Azimi, H.; Persaud, A.; Pyrsopoulos, N.T. Management and Treatment of Hepatocellular Carcinoma with Immunotherapy: A Review of Current and Future Options. J. Clin. Transl. Hepatol. 2020, 8, 168–176. [Google Scholar] [CrossRef]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.F.; De Oliveira, A.C.; Santoro, A.; Raoul, J.-L.; Forner, A.J.; et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef]
- Cheng, A.-L.; Kang, Y.-K.; Chen, Z.; Tsao, C.-J.; Qin, S.; Kim, J.S.; Luo, R.; Feng, J.; Ye, S.; Yang, T.-S.; et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: A phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2008, 10, 25–34. [Google Scholar] [CrossRef]
- Gozuacik, D.; Kimchi, A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene 2004, 23, 2891–2906. [Google Scholar] [CrossRef]
- Shimizu, S.; Takehara, T.; Hikita, H.; Kodama, T.; Tsunematsu, H.; Miyagi, T.; Hosui, A.; Ishida, H.; Tatsumi, T.; Kanto, T.; et al. Inhibition of autophagy potentiates the antitumor effect of the multikinase inhibitor sorafenib in hepatocellular carcinoma. Int. J. Cancer 2011, 131, 548–557. [Google Scholar] [CrossRef]
- Ringelhan, M.; Pfister, D.; O’Connor, T.; Pikarsky, E.; Heikenwalder, M. The immunology of hepatocellular carcinoma. Nat. Immunol. 2018, 19, 222–232. [Google Scholar] [CrossRef]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.-Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef]
- Yau, T.; Kang, Y.-K..; Kim, T.-Y.; El-Khoueiry, A.B.; Santoro, A.; Sangro, B.; Melero, I.; Kudo, M.; Hou, M.M.; Matilla, A.; et al. Efficacy and Safety of Nivolumab Plus Ipilimumab in Patients with Advanced Hepatocellular Carcinoma Previously Treated With Sorafenib: The CheckMate 040 Randomized Clinical Trial. JAMA Oncol. 2020, 6, e204564. [Google Scholar] [CrossRef]
- Jo, Y.; Kim, E.H.; Sai, S.; Kim, J.S.; Cho, J.-M.; Kim, H.; Baek, J.-H.; Kim, J.-Y.; Hwang, S.-G.; Yoon, M. Functional Biological Activity of Sorafenib as a Tumor-Treating Field Sensitizer for Glioblastoma Therapy. Int. J. Mol. Sci. 2018, 19, 3684. [Google Scholar] [CrossRef]
- Giladi, M.; Schneiderman, R.S.; Voloshin, T.; Porat, Y.; Munster, M.; Blat, R.; Sherbo, S.; Bomzon, Z.; Urman, N.; Itzhaki, A.; et al. Mitotic Spindle Disruption by Alternating Electric Fields Leads to Improper Chromosome Segregation and Mitotic Catastrophe in Cancer Cells. Sci. Rep. 2015, 5, 18046. [Google Scholar] [CrossRef]
- Kirson, E.D.; Dbalý, V.; Tovaryš, F.; Vymazal, J.; Soustiel, J.F.; Itzhaki, A.; Mordechovich, D.; Steinberg-Shapira, S.; Gurvich, Z.; Schneiderman, R.; et al. Alternating electric fields arrest cell proliferation in animal tumor models and human brain tumors. Proc. Natl. Acad. Sci. USA 2007, 104, 10152–10157. [Google Scholar] [CrossRef]
- Kirson, E.D.; Gurvich, Z.; Schneiderman, R.; Dekel, E.; Itzhaki, A.; Wasserman, Y.; Schatzberger, R.; Palti, Y. Disruption of Cancer Cell Replication by Alternating Electric Fields. Cancer Res. 2004, 64, 3288–3295. [Google Scholar] [CrossRef]
- Kim, E.H.; Kim, Y.J.; Song, H.S.; Jeong, Y.K.; Lee, J.Y.; Sung, J.; Yoo, S.H.; Yoon, M. Biological effect of an alternating electric field on cell proliferation and synergistic antimitotic effect in combination with ionizing radiation. Oncotarget 2016, 7, 62267–62279. [Google Scholar] [CrossRef]
- Karanam, N.K.; Ding, L.; Aroumougame, A.; Story, M.D. Tumor treating fields cause replication stress and interfere with DNA replication fork maintenance: Implications for cancer therapy. Transl. Res. 2020, 217, 33–46. [Google Scholar] [CrossRef]
- Karanam, N.K.; Srinivasan, K.; Ding, L.; Sishc, B.; Saha, D.; Story, M.D. Tumor-treating fields elicit a conditional vulnerability to ionizing radiation via the downregulation of BRCA1 signaling and reduced DNA double-strand break repair capacity in non-small cell lung cancer cell lines. Cell Death Dis. 2017, 8, e2711. [Google Scholar] [CrossRef]
- Giladi, M.; Munster, M.; Schneiderman, R.S.; Voloshin, T.; Porat, Y.; Blat, R.; Zielinska-Chomej, K.; Hååg, P.; Bomzon, Z.; Kirson, E.D.; et al. Tumor treating fields (TTFields) delay DNA damage repair following radiation treatment of glioma cells. Radiat. Oncol. 2017, 12, 206. [Google Scholar] [CrossRef]
- Mumblat, H.; Martinez-Conde, A.; Braten, O.; Munster, M.; Dor-On, E.; Schneiderman, R.S.; Porat, Y.; Voloshin, T.; Davidi, S.; Blatt, R.; et al. Tumor Treating Fields (TTFields) downregulate the Fanconi Anemia-BRCA pathway and increase the efficacy of chemotherapy in malignant pleural mesothelioma preclinical models. Lung Cancer 2021, 160, 99–110. [Google Scholar] [CrossRef]
- Shteingauz, A.; Porat, Y.; Voloshin, T.; Schneiderman, R.S.; Munster, M.; Zeevi, E.; Kaynan, N.; Gotlib, K.; Giladi, M.; Kirson, E.D.; et al. AMPK-dependent autophagy upregulation serves as a survival mechanism in response to Tumor Treating Fields (TTFields). Cell Death Dis. 2018, 9, 1074. [Google Scholar] [CrossRef]
- Voloshin, T.; Kaynan, N.; Davidi, S.; Porat, Y.; Shteingauz, A.; Schneiderman, R.S.; Zeevi, E.; Munster, M.; Blat, R.; Brami, C.T.; et al. Tumor-treating fields (TTFields) induce immunogenic cell death resulting in enhanced antitumor efficacy when combined with anti-PD-1 therapy. Cancer Immunol. Immunother. 2020, 69, 1191–1204. [Google Scholar] [CrossRef]
- Blatt, R.; Davidi, S.; Munster, M.; Shteingauz, A.; Cahal, S.; Zeidan, A.; Marciano, T.; Bomzon, Z.; Haber, A.; Giladi, M.; et al. In Vivo Safety of Tumor Treating Fields (TTFields) Applied to the Torso. Front. Oncol. 2021, 11, 670809. [Google Scholar] [CrossRef]
- Kanner, A.A.; Wong, E.T.; Villano, J.L.; Ram, Z. Post Hoc Analyses of Intention-to-Treat Population in Phase III Comparison of NovoTTF-100A™ System Versus Best Physician’s Choice Chemotherapy. Semin. Oncol. 2014, 41, S25–S34. [Google Scholar] [CrossRef]
- Toms, S.A.; Kim, C.Y.; Nicholas, G.; Ram, Z. Increased compliance with tumor treating fields therapy is prognostic for improved survival in the treatment of glioblastoma: A subgroup analysis of the EF-14 phase III trial. J. Neurooncol. 2018, 141, 467–473. [Google Scholar] [CrossRef]
- Hershkovich, H.S.; Urman, N.; Yesharim, O.; Naveh, A.; Bomzon, Z. The dielectric properties of skin and their influence on the delivery of tumor treating fields to the torso: A study combining in vivo measurements with numerical simulations. Phys. Med. Biol. 2019, 64, 185014. [Google Scholar] [CrossRef]
- Gera, N.; Yang, A.; Holtzman, T.S.; Lee, S.X.; Wong, E.T.; Swanson, K.D. Tumor Treating Fields Perturb the Localization of Septins and Cause Aberrant Mitotic Exit. PLoS ONE 2015, 10, e0125269. [Google Scholar] [CrossRef]
- Mauthe, M.; Orhon, I.; Rocchi, C.; Zhou, X.; Luhr, M.; Hijlkema, K.-J.; Coppes, R.P.; Engedal, N.; Mari, M.; Reggiori, F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 2018, 14, 1435–1455. [Google Scholar] [CrossRef]
- Rodríguez-Hernández, M.A.; González, R.; de la Rosa, Á.J.; Gallego, P.; Ordóñez, R.; Navarro-Villarán, E.; Contreras, L.; Rodríguez-Arribas, M.; González-Gallego, J.; Álamo-Martínez, J.M.; et al. Molecular characterization of autophagic and apoptotic signaling induced by sorafenib in liver cancer cells. J. Cell. Physiol. 2018, 234, 692–708. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, G.; Chen, Y.; Wang, H.; Hua, Y.; Cai, Z. Immunogenic cell death in cancer therapy: Present and emerging inducers. J. Cell. Mol. Med. 2019, 23, 4854–4865. [Google Scholar] [CrossRef] [PubMed]
- Silginer, M.; Weller, M.; Stupp, R.; Roth, P. Biological activity of tumor-treating fields in preclinical glioma models. Cell Death Dis. 2017, 8, e2753. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.H.; Jo, Y.; Sai, S.; Park, M.J.; Kim, J.Y.; Kim, J.S.; Lee, Y.-J.; Cho, J.-M.; Kwak, S.-Y.; Baek, J.-H.; et al. Tumor-treating fields induce autophagy by blocking the Akt2/miR29b axis in glioblastoma cells. Oncogene 2019, 38, 6630–6646. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Toscano, A.; Nickel, A.-C.; Li, G.; Kamp, M.A.; Muhammad, S.; Leprivier, G.; Fritsche, E.; Barker, R.A.; Sabel, M.; Steiger, H.-J.; et al. Rapalink-1 Targets Glioblastoma Stem Cells and Acts Synergistically with Tumor Treating Fields to Reduce Resistance against Temozolomide. Cancers 2020, 12, 3859. [Google Scholar] [CrossRef]
- Gkika, E.; Grosu, A.-L.; Mercade, T.M.; Gracián, A.C.; Brunner, T.B.; Schultheiß, M.; Pazgan-Simon, M.; Seufferlein, T.; Touchefeu, Y. Tumor Treating Fields Concomitant with Sorafenib in Advanced Hepatocellular Cancer: Results of the HEPANOVA Phase II Study. Cancers 2022, 14, 1568. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davidi, S.; Jacobovitch, S.; Shteingauz, A.; Martinez-Conde, A.; Braten, O.; Tempel-Brami, C.; Zeevi, E.; Frechtel-Gerzi, R.; Ene, H.; Dor-On, E.; et al. Tumor Treating Fields (TTFields) Concomitant with Sorafenib Inhibit Hepatocellular Carcinoma In Vitro and In Vivo. Cancers 2022, 14, 2959. https://doi.org/10.3390/cancers14122959
Davidi S, Jacobovitch S, Shteingauz A, Martinez-Conde A, Braten O, Tempel-Brami C, Zeevi E, Frechtel-Gerzi R, Ene H, Dor-On E, et al. Tumor Treating Fields (TTFields) Concomitant with Sorafenib Inhibit Hepatocellular Carcinoma In Vitro and In Vivo. Cancers. 2022; 14(12):2959. https://doi.org/10.3390/cancers14122959
Chicago/Turabian StyleDavidi, Shiri, Sara Jacobovitch, Anna Shteingauz, Antonia Martinez-Conde, Ori Braten, Catherine Tempel-Brami, Einav Zeevi, Roni Frechtel-Gerzi, Hila Ene, Eyal Dor-On, and et al. 2022. "Tumor Treating Fields (TTFields) Concomitant with Sorafenib Inhibit Hepatocellular Carcinoma In Vitro and In Vivo" Cancers 14, no. 12: 2959. https://doi.org/10.3390/cancers14122959





