Insights of RKIP-Derived Suppression of Prostate Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. The Molecular Basis for RKIP as a Tumor Suppressor
3. RKIP as a Tumor Suppression of Prostate Cancer (PC)
3.1. RKIP-Mediated Suppression of PC Tumorigenesis and Metastasis
3.1.1. Facilitation of PC Initiation and Metastasis via Downregulation of RKIP at the Protein Level
3.1.2. No Apparent Reduction of RKIP mRNA Expression Following PC Pathogenesis
3.2. Regulation of RKIP Expression in PC Cells
3.2.1. RKIP as a Target of Androgen Receptor (AR)
3.2.2. Mutual Regulation of RKIP and the EMT Machinery
3.2.3. Non-Coding RNA-Mediated Downregulation of RKIP in PC Cells
3.3. RKIP-Derived Sensitization of PC Cells to Treatment In Vitro
4. Utilization of Additional Mechanisms in RKIP-Derived Inhibition of PC
4.1. A Potential Interplay between Two PC Metastasis Suppressors: RKIP and Annexin A7
4.2. EZH2-Derived Downregulation of RKIP Transcription in PC
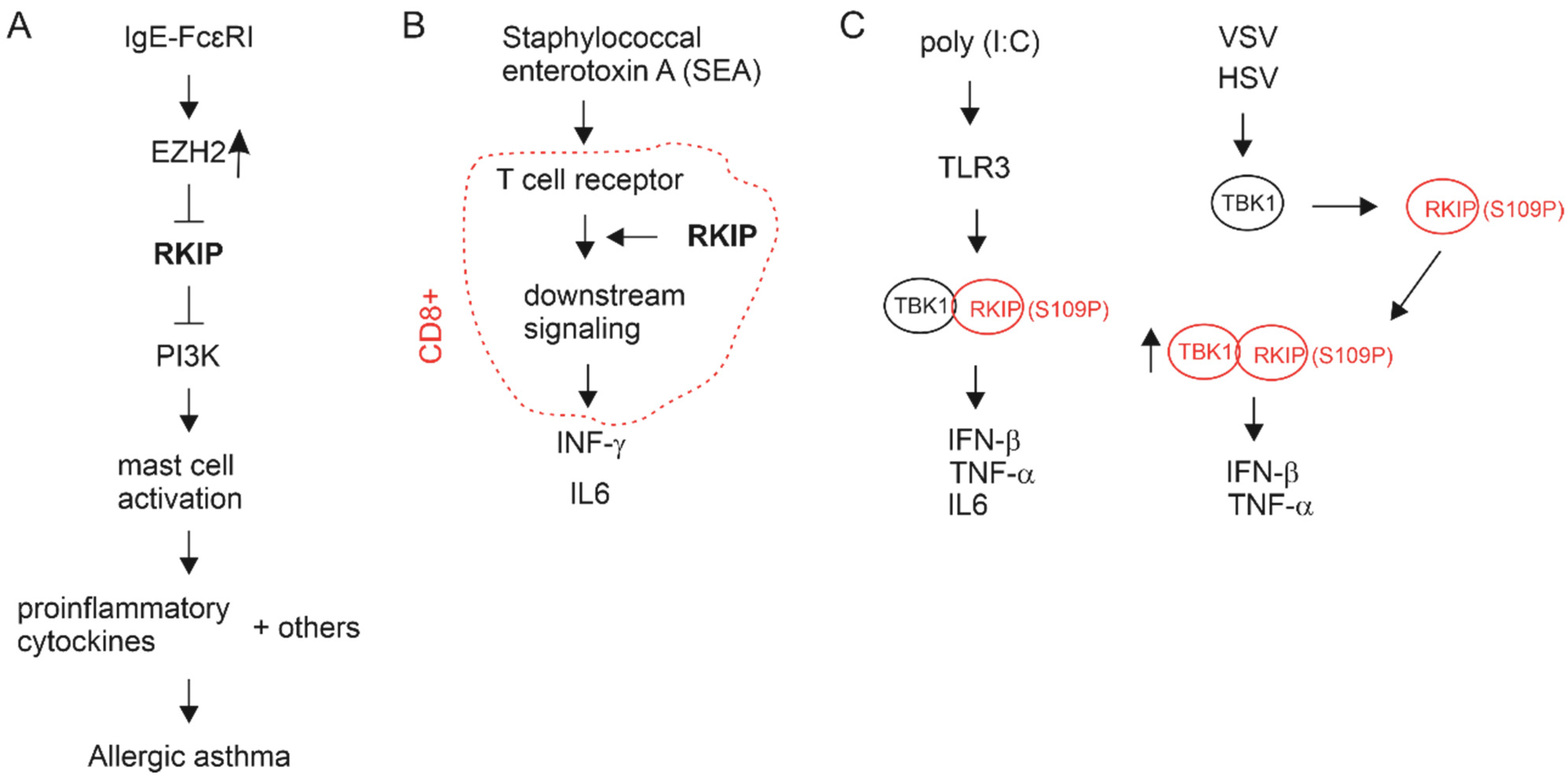
4.3. Immune Alterations Associated with RKIP in PC
5. Perspectives
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Heidenreich, A.; Bastian, P.J.; Bellmunt, J.; Bolla, M.; Joniau, S.; van der Kwast, T.; Mason, M.; Matveev, V.; Wiegel, T.; Zattoni, F.; et al. EAU guidelines on prostate cancer. Part II: Treatment of advanced, relapsing, and castration-resistant prostate cancer. Eur. Urol. 2014, 65, 467–479. [Google Scholar] [CrossRef]
- Egevad, L.; Delahunt, B.; Srigley, J.R.; Samaratunga, H. International Society of Urological Pathology (ISUP) grading of prostate cancer—An ISUP consensus on contemporary grading. APMIS Acta Pathol. Microbiol. Immunol. Scand. 2016, 124, 433–435. [Google Scholar] [CrossRef] [PubMed]
- Gordetsky, J.; Epstein, J. Grading of prostatic adenocarcinoma: Current state and prognostic implications. Diagn. Pathol. 2016, 11, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epstein, J.I.; Zelefsky, M.J.; Sjoberg, D.D.; Nelson, J.B.; Egevad, L.; Magi-Galluzzi, C.; Vickers, A.J.; Parwani, A.V.; Reuter, V.E.; Fine, S.W.; et al. A Contemporary Prostate Cancer Grading System: A Validated Alternative to the Gleason Score. Eur. Urol. 2016, 69, 428–435. [Google Scholar] [CrossRef] [Green Version]
- Mottet, N.; Bellmunt, J.; Bolla, M.; Briers, E.; Cumberbatch, M.G.; De Santis, M.; Fossati, N.; Gross, T.; Henry, A.M.; Joniau, S.; et al. EAU-ESTRO-SIOG Guidelines on Prostate Cancer. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2017, 71, 618–629. [Google Scholar] [CrossRef]
- Bill-Axelson, A.; Holmberg, L.; Garmo, H.; Rider, J.R.; Taari, K.; Busch, C.; Nordling, S.; Haggman, M.; Andersson, S.O.; Spangberg, A.; et al. Radical prostatectomy or watchful waiting in early prostate cancer. N. Engl. J. Med. 2014, 370, 932–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayes, J.H.; Ollendorf, D.A.; Pearson, S.D.; Barry, M.J.; Kantoff, P.W.; Lee, P.A.; McMahon, P.M. Observation versus initial treatment for men with localized, low-risk prostate cancer: A cost-effectiveness analysis. Ann. Intern. Med. 2013, 158, 853–860. [Google Scholar] [CrossRef]
- Godtman, R.A.; Holmberg, E.; Khatami, A.; Stranne, J.; Hugosson, J. Outcome following active surveillance of men with screen-detected prostate cancer. Results from the Goteborg randomised population-based prostate cancer screening trial. Eur. Urol. 2013, 63, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Zaorsky, N.G.; Raj, G.V.; Trabulsi, E.J.; Lin, J.; Den, R.B. The dilemma of a rising prostate-specific antigen level after local therapy: What are our options? Semin. Oncol. 2013, 40, 322–336. [Google Scholar] [CrossRef] [Green Version]
- Cornford, P.; Bellmunt, J.; Bolla, M.; Briers, E.; De Santis, M.; Gross, T.; Henry, A.M.; Joniau, S.; Lam, T.B.; Mason, M.D.; et al. EAU-ESTRO-SIOG Guidelines on Prostate Cancer. Part II: Treatment of Relapsing, Metastatic, and Castration-Resistant Prostate Cancer. Eur. Urol. 2017, 71, 630–642. [Google Scholar] [CrossRef]
- Pound, C.R.; Partin, A.W.; Eisenberger, M.A.; Chan, D.W.; Pearson, J.D.; Walsh, P.C. Natural history of progression after PSA elevation following radical prostatectomy. JAMA 1999, 281, 1591–1597. [Google Scholar] [CrossRef] [Green Version]
- Boorjian, S.A.; Thompson, R.H.; Tollefson, M.K.; Rangel, L.J.; Bergstralh, E.J.; Blute, M.L.; Karnes, R.J. Long-term risk of clinical progression after biochemical recurrence following radical prostatectomy: The impact of time from surgery to recurrence. Eur. Urol. 2011, 59, 893–899. [Google Scholar] [CrossRef]
- Lytton, B. Prostate cancer: A brief history and the discovery of hormonal ablation treatment. J. Urol 2001, 165, 1859–1862. [Google Scholar] [CrossRef]
- Wong, Y.N.; Ferraldeschi, R.; Attard, G.; de Bono, J. Evolution of androgen receptor targeted therapy for advanced prostate cancer. Nat. Rev. Clin. Oncol. 2014, 11, 365–376. [Google Scholar] [CrossRef]
- Semenas, J.; Allegrucci, C.; Boorjian, S.A.; Mongan, N.P.; Persson, J.L. Overcoming drug resistance and treating advanced prostate cancer. Curr. Drug Targets 2012, 13, 1308–1323. [Google Scholar] [CrossRef] [Green Version]
- Ojo, D.; Lin, X.; Wong, N.; Gu, Y.; Tang, D. Prostate Cancer Stem-like Cells Contribute to the Development of Castration-Resistant Prostate Cancer. Cancers 2015, 7, 2290–2308. [Google Scholar] [CrossRef]
- de Bono, J.S.; Logothetis, C.J.; Molina, A.; Fizazi, K.; North, S.; Chu, L.; Chi, K.N.; Jones, R.J.; Goodman, O.B., Jr.; Saad, F.; et al. Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 2011, 364, 1995–2005. [Google Scholar] [CrossRef]
- Scher, H.I.; Fizazi, K.; Saad, F.; Taplin, M.E.; Sternberg, C.N.; Miller, K.; de Wit, R.; Mulders, P.; Chi, K.N.; Shore, N.D.; et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 2012, 367, 1187–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaturvedi, S.; Garcia, J.A. Novel agents in the management of castration resistant prostate cancer. J. Carcinog. 2014, 13, 5. [Google Scholar] [CrossRef] [PubMed]
- Drake, C.G. Prostate cancer as a model for tumour immunotherapy. Nat. Rev. Immunol. 2010, 10, 580–593. [Google Scholar] [CrossRef] [Green Version]
- Mei, W.; Gu, Y.; Jiang, Y.; Major, P.; Tang, D. Circulating cell-free DNA is a potential prognostic biomarker of metastatic castration-resistant prostate cancer for taxane therapy. AME Med. J. 2018, 3, 1–5. [Google Scholar] [CrossRef]
- Weigelt, B.; Peterse, J.L.; van ‘t Veer, L.J. Breast cancer metastasis: Markers and models. Nat. Rev. Cancer 2005, 5, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Gupta, G.P.; Massague, J. Cancer metastasis: Building a framework. Cell 2006, 127, 679–695. [Google Scholar] [CrossRef] [Green Version]
- De Craene, B.; Berx, G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 2013, 13, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.H.; Yang, J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013, 27, 2192–2206. [Google Scholar] [CrossRef] [Green Version]
- Shibue, T.; Weinberg, R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 611–629. [Google Scholar] [CrossRef] [Green Version]
- Mei, W.; Lin, X.; Kapoor, A.; Gu, Y.; Zhao, K.; Tang, D. The Contributions of Prostate Cancer Stem Cells in Prostate Cancer Initiation and Metastasis. Cancers 2019, 11, 434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, C.; Theodorescu, D. RAL GTPases: Biology and Potential as Therapeutic Targets in Cancer. Pharmacol. Rev. 2018, 70, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Makwana, V.; Rudrawar, S.; Anoopkumar-Dukie, S. Signalling transduction of O-GlcNAcylation and PI3K/AKT/mTOR-axis in prostate cancer. Biochim. Biophys. Acta. Mol. Basis Dis. 2021, 1867, 166129. [Google Scholar] [CrossRef]
- Wu, X.; Scott, H.; Carlsson, S.V.; Sjoberg, D.D.; Cerundolo, L.; Lilja, H.; Prevo, R.; Rieunier, G.; Macaulay, V.; Higgins, G.S.; et al. Increased EZH2 expression in prostate cancer is associated with metastatic recurrence following external beam radiotherapy. Prostate 2019, 79, 1079–1089. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Wang, G.; Li, Q.; Jiang, W.; Kim, J.S.; Wang, R.; Zhu, S.; Wang, X.; Yan, L.; Yi, Y.; et al. Polycomb group proteins EZH2 and EED directly regulate androgen receptor in advanced prostate cancer. Int. J. Cancer 2019, 145, 415–426. [Google Scholar] [CrossRef]
- Zhao, Y.; Ding, L.; Wang, D.; Ye, Z.; He, Y.; Ma, L.; Zhu, R.; Pan, Y.; Wu, Q.; Pang, K.; et al. EZH2 cooperates with gain-of-function p53 mutants to promote cancer growth and metastasis. EMBO J. 2019, 38, e99599. [Google Scholar] [CrossRef]
- Ren, D.; Yang, Q.; Dai, Y.; Guo, W.; Du, H.; Song, L.; Peng, X. Oncogenic miR-210-3p promotes prostate cancer cell EMT and bone metastasis via NF-kappaB signaling pathway. Mol. Cancer 2017, 16, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaltschmidt, C.; Banz-Jansen, C.; Benhidjeb, T.; Beshay, M.; Forster, C.; Greiner, J.; Hamelmann, E.; Jorch, N.; Mertzlufft, F.; Pfitzenmaier, J.; et al. A Role for NF-kappaB in Organ Specific Cancer and Cancer Stem Cells. Cancers 2019, 11, 655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller, E.T.; Fu, Z.; Brennan, M. The biology of a prostate cancer metastasis suppressor protein: Raf kinase inhibitor protein. J. Cell Biochem. 2005, 94, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Keller, E.T.; Fu, Z.; Yeung, K.; Brennan, M. Raf kinase inhibitor protein: A prostate cancer metastasis suppressor gene. Cancer Lett. 2004, 207, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Bernier, I.; Jolles, P. Purification and characterization of a basic 23 kDa cytosolic protein from bovine brain. Biochim. Biophys. Acta 1984, 790, 174–181. [Google Scholar] [CrossRef]
- Hori, N.; Chae, K.S.; Murakawa, K.; Matoba, R.; Fukushima, A.; Okubo, K.; Matsubara, K. A human cDNA sequence homologue of bovine phosphatidylethanolamine-binding protein. Gene 1994, 140, 293–294. [Google Scholar] [CrossRef] [PubMed]
- Seddiqi, N.; Bollengier, F.; Alliel, P.M.; Perin, J.P.; Bonnet, F.; Bucquoy, S.; Jolles, P.; Schoentgen, F. Amino acid sequence of the Homo sapiens brain 21-23-kDa protein (neuropolypeptide h3), comparison with its counterparts from Rattus norvegicus and Bos taurus species, and expression of its mRNA in different tissues. J. Mol. Evol. 1994, 39, 655–660. [Google Scholar] [CrossRef]
- Perry, A.C.; Hall, L.; Bell, A.E.; Jones, R. Sequence analysis of a mammalian phospholipid-binding protein from testis and epididymis and its distribution between spermatozoa and extracellular secretions. Biochem. J. 1994, 301 Pt 1, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Grandy, D.K.; Hanneman, E.; Bunzow, J.; Shih, M.; Machida, C.A.; Bidlack, J.M.; Civelli, O. Purification, cloning, and tissue distribution of a 23-kDa rat protein isolated by morphine affinity chromatography. Mol. Endocrinol. 1990, 4, 1370–1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araki, Y.; Vierula, M.E.; Rankin, T.L.; Tulsiani, D.R.; Orgebin-Crist, M.C. Isolation and characterization of a 25-kilodalton protein from mouse testis: Sequence homology with a phospholipid-binding protein. Biol. Reprod. 1992, 47, 832–843. [Google Scholar] [CrossRef] [Green Version]
- Bollengier, F.; Mahler, A. Localization of the novel neuropolypeptide h3 in subsets of tissues from different species. J. Neurochem. 1988, 50, 1210–1214. [Google Scholar] [CrossRef] [PubMed]
- Schoentgen, F.; Jolles, P. From structure to function: Possible biological roles of a new widespread protein family binding hydrophobic ligands and displaying a nucleotide binding site. FEBS Lett. 1995, 369, 22–26. [Google Scholar] [CrossRef] [Green Version]
- Pikielny, C.W.; Hasan, G.; Rouyer, F.; Rosbash, M. Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron 1994, 12, 35–49. [Google Scholar] [CrossRef]
- Robinson, L.C.; Tatchell, K. TFS1: A suppressor of cdc25 mutations in Saccharomyces cerevisiae. Mol. Gen. Genet. 1991, 230, 241–250. [Google Scholar] [CrossRef]
- Gems, D.; Ferguson, C.J.; Robertson, B.D.; Nieves, R.; Page, A.P.; Blaxter, M.L.; Maizels, R.M. An abundant, trans-spliced mRNA from Toxocara canis infective larvae encodes a 26-kDa protein with homology to phosphatidylethanolamine-binding proteins. J. Biol. Chem. 1995, 270, 18517–18522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erttmann, K.D.; Gallin, M.Y. Onchocerca volvulus: Identification of cDNAs encoding a putative phosphatidyl-ethanolamine-binding protein and a putative partially processed mRNA precursor. Gene 1996, 174, 203–207. [Google Scholar] [CrossRef]
- Trottein, F.; Cowman, A.F. The primary structure of a putative phosphatidylethanolamine-binding protein from Plasmodium falciparum. Mol. Biochem. Parasitol. 1995, 70, 235–239. [Google Scholar] [CrossRef]
- Bradley, D.; Carpenter, R.; Copsey, L.; Vincent, C.; Rothstein, S.; Coen, E. Control of inflorescence architecture in Antirrhinum. Nature 1996, 379, 791–797. [Google Scholar] [CrossRef]
- Ohshima, S.; Murata, M.; Sakamoto, W.; Ogura, Y.; Motoyoshi, F. Cloning and molecular analysis of the Arabidopsis gene Terminal Flower 1. Mol. Gen. Genet. 1997, 254, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Yeung, K.; Seitz, T.; Li, S.; Janosch, P.; McFerran, B.; Kaiser, C.; Fee, F.; Katsanakis, K.D.; Rose, D.W.; Mischak, H.; et al. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 1999, 401, 173–177. [Google Scholar] [CrossRef] [Green Version]
- Corbit, K.C.; Trakul, N.; Eves, E.M.; Diaz, B.; Marshall, M.; Rosner, M.R. Activation of Raf-1 signaling by protein kinase C through a mechanism involving Raf kinase inhibitory protein. J. Biol. Chem. 2003, 278, 13061–13068. [Google Scholar] [CrossRef] [Green Version]
- Yotova, I.; Quan, P.; Gaba, A.; Leditznig, N.; Pateisky, P.; Kurz, C.; Tschugguel, W. Raf-1 levels determine the migration rate of primary endometrial stromal cells of patients with endometriosis. J. Cell Mol. Med. 2012, 16, 2127–2139. [Google Scholar] [CrossRef]
- Evron, T.; Daigle, T.L.; Caron, M.G. GRK2: Multiple roles beyond G protein-coupled receptor desensitization. Trends Pharmacol. Sci. 2012, 33, 154–164. [Google Scholar] [CrossRef] [Green Version]
- Lorenz, K.; Lohse, M.J.; Quitterer, U. Protein kinase C switches the Raf kinase inhibitor from Raf-1 to GRK-2. Nature 2003, 426, 574–579. [Google Scholar] [CrossRef] [PubMed]
- Schmid, E.; Neef, S.; Berlin, C.; Tomasovic, A.; Kahlert, K.; Nordbeck, P.; Deiss, K.; Denzinger, S.; Herrmann, S.; Wettwer, E.; et al. Cardiac RKIP induces a beneficial beta-adrenoceptor-dependent positive inotropy. Nat. Med. 2015, 21, 1298–1306. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, K.; Rosner, M.R.; Brand, T.; Schmitt, J.P. Raf kinase inhibitor protein: Lessons of a better way for beta-adrenergic receptor activation in the heart. J. Physiol. 2017, 595, 4073–4087. [Google Scholar] [CrossRef]
- Degirmenci, U.; Wang, M.; Hu, J. Targeting Aberrant RAS/RAF/MEK/ERK Signaling for Cancer Therapy. Cells 2020, 9, 198. [Google Scholar] [CrossRef] [Green Version]
- Asati, V.; Mahapatra, D.K.; Bharti, S.K. PI3K/Akt/mTOR and Ras/Raf/MEK/ERK signaling pathways inhibitors as anticancer agents: Structural and pharmacological perspectives. Eur. J. Med. Chem. 2016, 109, 314–341. [Google Scholar] [CrossRef] [PubMed]
- Roberts, P.J.; Der, C.J. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 2007, 26, 3291–3310. [Google Scholar] [CrossRef] [Green Version]
- Dorsam, R.T.; Gutkind, J.S. G-protein-coupled receptors and cancer. Nat. Rev. Cancer 2007, 7, 79–94. [Google Scholar] [CrossRef]
- Bar-Shavit, R.; Maoz, M.; Kancharla, A.; Nag, J.K.; Agranovich, D.; Grisaru-Granovsky, S.; Uziely, B. G Protein-Coupled Receptors in Cancer. Int. J. Mol. Sci. 2016, 17, 1320. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, H.; Hagerling, C.; Werb, Z. Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes Dev. 2018, 32, 1267–1284. [Google Scholar] [CrossRef] [Green Version]
- Humbert, M.; Bousquet, J.; Bachert, C.; Palomares, O.; Pfister, P.; Kottakis, I.; Jaumont, X.; Thomsen, S.F.; Papadopoulos, N.G. IgE-Mediated Multimorbidities in Allergic Asthma and the Potential for Omalizumab Therapy. J. Allergy Clin. Immunol. Pract. 2019, 7, 1418–1429. [Google Scholar] [CrossRef]
- Lin, W.C.; Hsu, F.S.; Kuo, K.L.; Liu, S.H.; Shun, C.T.; Shi, C.S.; Chang, H.C.; Tsai, Y.C.; Lin, M.C.; Wu, J.T.; et al. Correction to: Trichostatin A, a histone deacetylase inhibitor, induces synergistic cytotoxicity with chemotherapy via suppression of Raf/MEK/ERK pathway in urothelial carcinoma. J. Mol. Med. 2019, 97, 435–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, N.; Baby, D.; Rajguru, J.P.; Patil, P.B.; Thakkannavar, S.S.; Pujari, V.B. Inflammation and cancer. Ann. Afr. Med. 2019, 18, 121–126. [Google Scholar] [CrossRef]
- Wright, K.T.; Vella, A.T. RKIP contributes to IFN-gamma synthesis by CD8+ T cells after serial TCR triggering in systemic inflammatory response syndrome. J. Immunol. 2013, 191, 708–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farhood, B.; Najafi, M.; Mortezaee, K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: A review. J. Cell. Physiol. 2019, 234, 8509–8521. [Google Scholar] [CrossRef]
- Lai, R.; Gu, M.; Jiang, W.; Lin, W.; Xu, P.; Liu, Z.; Huang, H.; An, H.; Wang, X. Raf Kinase Inhibitor Protein Preferentially Promotes TLR3-Triggered Signaling and Inflammation. J. Immunol. 2017, 198, 4086–4095. [Google Scholar] [CrossRef] [Green Version]
- Gu, M.; Liu, Z.; Lai, R.; Liu, S.; Lin, W.; Ouyang, C.; Ye, S.; Huang, H.; Wang, X. RKIP and TBK1 form a positive feedback loop to promote type I interferon production in innate immunity. EMBO J. 2016, 35, 2553–2565. [Google Scholar] [CrossRef] [PubMed]
- Ostuni, R.; Kratochvill, F.; Murray, P.J.; Natoli, G. Macrophages and cancer: From mechanisms to therapeutic implications. Trends Immunol. 2015, 36, 229–239. [Google Scholar] [CrossRef]
- Jorgovanovic, D.; Song, M.; Wang, L.; Zhang, Y. Roles of IFN-gamma in tumor progression and regression: A review. Biomark. Res. 2020, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Josephs, S.F.; Ichim, T.E.; Prince, S.M.; Kesari, S.; Marincola, F.M.; Escobedo, A.R.; Jafri, A. Unleashing endogenous TNF-alpha as a cancer immunotherapeutic. J. Transl. Med. 2018, 16, 242. [Google Scholar] [CrossRef] [PubMed]
- Snell, L.M.; McGaha, T.L.; Brooks, D.G. Type I Interferon in Chronic Virus Infection and Cancer. Trends Immunol. 2017, 38, 542–557. [Google Scholar] [CrossRef]
- Yeung, K.C.; Rose, D.W.; Dhillon, A.S.; Yaros, D.; Gustafsson, M.; Chatterjee, D.; McFerran, B.; Wyche, J.; Kolch, W.; Sedivy, J.M. Raf kinase inhibitor protein interacts with NF-kappaB-inducing kinase and TAK1 and inhibits NF-kappaB activation. Mol. Cell. Biol. 2001, 21, 7207–7217. [Google Scholar] [CrossRef] [Green Version]
- Hayden, M.S.; Ghosh, S. NF-kappaB in immunobiology. Cell Res. 2011, 21, 223–244. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.C. The non-canonical NF-kappaB pathway in immunity and inflammation. Nat. Rev. Immunol. 2017, 17, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, K.; Karin, M. NF-kappaB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 2018, 18, 309–324. [Google Scholar] [CrossRef]
- Al-Mulla, F.; Bitar, M.S.; Al-Maghrebi, M.; Behbehani, A.I.; Al-Ali, W.; Rath, O.; Doyle, B.; Tan, K.Y.; Pitt, A.; Kolch, W. Raf kinase inhibitor protein RKIP enhances signaling by glycogen synthase kinase-3beta. Cancer Res. 2011, 71, 1334–1343. [Google Scholar] [CrossRef] [Green Version]
- Thornton, T.M.; Pedraza-Alva, G.; Deng, B.; Wood, C.D.; Aronshtam, A.; Clements, J.L.; Sabio, G.; Davis, R.J.; Matthews, D.E.; Doble, B.; et al. Phosphorylation by p38 MAPK as an alternative pathway for GSK3beta inactivation. Science 2008, 320, 667–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fatima, I.; Singh, A.B.; Dhawan, P. MASTL: A novel therapeutic target for Cancer Malignancy. Cancer Med. 2020, 9, 6322–6329. [Google Scholar] [CrossRef]
- Zaravinos, A.; Chatziioannou, M.; Lambrou, G.I.; Boulalas, I.; Delakas, D.; Spandidos, D.A. Implication of RAF and RKIP genes in urinary bladder cancer. Pathol. Oncol. Res. POR 2011, 17, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Moon, A.; Park, J.Y.; Sung, J.Y.; Park, Y.K.; Kim, Y.W. Reduced expression of Raf-1 kinase inhibitory protein in renal cell carcinoma: A significant prognostic marker. Pathology 2012, 44, 534–539. [Google Scholar] [CrossRef]
- Hill, B.; De Melo, J.; Yan, J.; Kapoor, A.; He, L.; Cutz, J.C.; Feng, X.; Bakhtyar, N.; Tang, D. Common reduction of the Raf kinase inhibitory protein in clear cell renal cell carcinoma. Oncotarget 2014, 5, 7406–7419. [Google Scholar] [CrossRef] [PubMed]
- Papale, M.; Vocino, G.; Lucarelli, G.; Rutigliano, M.; Gigante, M.; Rocchetti, M.T.; Pesce, F.; Sanguedolce, F.; Bufo, P.; Battaglia, M.; et al. Urinary RKIP/p-RKIP is a potential diagnostic and prognostic marker of clear cell renal cell carcinoma. Oncotarget 2017, 8, 40412–40424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Z.; Smith, P.C.; Zhang, L.; Rubin, M.A.; Dunn, R.L.; Yao, Z.; Keller, E.T. Effects of raf kinase inhibitor protein expression on suppression of prostate cancer metastasis. Natl. Cancer Inst. 2003, 95, 878–889. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Kitagawa, Y.; Shen, R.; Shah, R.; Mehra, R.; Rhodes, D.; Keller, P.J.; Mizokami, A.; Dunn, R.; Chinnaiyan, A.M.; et al. Metastasis suppressor gene Raf kinase inhibitor protein (RKIP) is a novel prognostic marker in prostate cancer. Prostate 2006, 66, 248–256. [Google Scholar] [CrossRef] [Green Version]
- Dangi-Garimella, S.; Yun, J.; Eves, E.M.; Newman, M.; Erkeland, S.J.; Hammond, S.M.; Minn, A.J.; Rosner, M.R. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009, 28, 347–358. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, S.; Tang, X.; Zhang, A.; Grabinski, T.; Guo, Z.; Hudson, E.; Berghuis, B.; Webb, C.; Zhao, P.; et al. Development and evaluation of monoclonal antibodies against phosphatidylethanolamine binding protein 1 in pancreatic cancer patients. J. Immunol. Methods 2010, 362, 151–160. [Google Scholar] [CrossRef]
- Lee, H.C.; Tian, B.; Sedivy, J.M.; Wands, J.R.; Kim, M. Loss of Raf kinase inhibitor protein promotes cell proliferation and migration of human hepatoma cells. Gastroenterology 2006, 131, 1208–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Wu, X.; Wu, T.; Li, G.M.; Shi, Y. Clinical significance of RKIP mRNA expression in non-small cell lung cancer. Tumour. Biol. 2014, 35, 4377–4380. [Google Scholar] [CrossRef]
- Zhang, X.M.; Gu, H.; Yan, L.; Zhang, G.Y. RKIP inhibits the malignant phenotypes of gastric cancer cells. Neoplasma 2013, 60, 196–202. [Google Scholar] [CrossRef] [Green Version]
- Lamiman, K.; Keller, J.M.; Mizokami, A.; Zhang, J.; Keller, E.T. Survey of Raf kinase inhibitor protein (RKIP) in multiple cancer types. Crit. Rev. Oncog. 2014, 19, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Dozmorov, I.M.; Keller, E.T. Osteoblasts produce soluble factors that induce a gene expression pattern in non-metastatic prostate cancer cells, similar to that found in bone metastatic prostate cancer cells. Prostate 2002, 51, 10–20. [Google Scholar] [CrossRef]
- Escara-Wilke, J.; Keller, J.M.; Ignatoski, K.M.; Dai, J.; Shelley, G.; Mizokami, A.; Zhang, J.; Yeung, M.L.; Yeung, K.C.; Keller, E.T. Raf kinase inhibitor protein (RKIP) deficiency decreases latency of tumorigenesis and increases metastasis in a murine genetic model of prostate cancer. Prostate 2015, 75, 292–302. [Google Scholar] [CrossRef] [Green Version]
- Theroux, S.; Pereira, M.; Casten, K.S.; Burwell, R.D.; Yeung, K.C.; Sedivy, J.M.; Klysik, J. Raf kinase inhibitory protein knockout mice: Expression in the brain and olfaction deficit. Brain Res. Bull. 2007, 71, 559–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [Green Version]
- Taylor, B.S.; Schultz, N.; Hieronymus, H.; Gopalan, A.; Xiao, Y.; Carver, B.S.; Arora, V.K.; Kaushik, P.; Cerami, E.; Reva, B.; et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010, 18, 11–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsiades, N. A road map to comprehensive androgen receptor axis targeting for castration-resistant prostate cancer. Cancer Res. 2013, 73, 4599–4605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mateo, J.; Smith, A.; Ong, M.; de Bono, J.S. Novel drugs targeting the androgen receptor pathway in prostate cancer. Cancer Metastasis Rev. 2014, 33, 567–579. [Google Scholar] [CrossRef]
- Bello, D.; Webber, M.M.; Kleinman, H.K.; Wartinger, D.D.; Rhim, J.S. Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis 1997, 18, 1215–1223. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, J.; Keller, J.M.; Yeung, K.; Keller, E.T.; Fu, Z. Transcriptional regulation of RKIP expression by androgen in prostate cells. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2012, 30, 1340–1350. [Google Scholar] [CrossRef]
- Yeh, S.; Tsai, M.Y.; Xu, Q.; Mu, X.M.; Lardy, H.; Huang, K.E.; Lin, H.; Yeh, S.D.; Altuwaijri, S.; Zhou, X.; et al. Generation and characterization of androgen receptor knockout (ARKO) mice: An in vivo model for the study of androgen functions in selective tissues. Proc. Natl. Acad. Sci. USA 2002, 99, 13498–13503. [Google Scholar] [CrossRef] [Green Version]
- Quigley, C.A.; De Bellis, A.; Marschke, K.B.; el-Awady, M.K.; Wilson, E.M.; French, F.S. Androgen receptor defects: Historical, clinical, and molecular perspectives. Endocr. Rev. 1995, 16, 271–321. [Google Scholar] [CrossRef]
- Heinlein, C.A.; Chang, C. Androgen receptor in prostate cancer. Endocr. Rev. 2004, 25, 276–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Coetzee, G.A. Prostate specific antigen gene regulation by androgen receptor. J. Cell Biochem. 2004, 93, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Ben Jemaa, A.; Bouraoui, Y.; Sallami, S.; Nouira, Y.; Oueslati, R. A comparison of the biological features of prostate cancer with (PSA+, PSMA+) profile according to RKIP. Biomed. Res. Int. 2013, 2013, 409179. [Google Scholar] [CrossRef] [PubMed]
- Roehl, K.A.; Han, M.; Ramos, C.G.; Antenor, J.A.; Catalona, W.J. Cancer progression and survival rates following anatomical radical retropubic prostatectomy in 3478 consecutive patients: Long-term results. J. Urol. 2004, 172, 910–914. [Google Scholar] [CrossRef] [Green Version]
- Freedland, S.J.; Humphreys, E.B.; Mangold, L.A.; Eisenberger, M.; Dorey, F.J.; Walsh, P.C.; Partin, A.W. Risk of prostate cancer-specific mortality following biochemical recurrence after radical prostatectomy. JAMA 2005, 294, 433–439. [Google Scholar] [CrossRef] [Green Version]
- Verras, M.; Lee, J.; Xue, H.; Li, T.H.; Wang, Y.; Sun, Z. The androgen receptor negatively regulates the expression of c-Met: Implications for a novel mechanism of prostate cancer progression. Cancer Res. 2007, 67, 967–975. [Google Scholar] [CrossRef] [Green Version]
- Carver, B.S.; Chapinski, C.; Wongvipat, J.; Hieronymus, H.; Chen, Y.; Chandarlapaty, S.; Arora, V.K.; Le, C.; Koutcher, J.; Scher, H.; et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 2011, 19, 575–586. [Google Scholar] [CrossRef] [Green Version]
- Mulholland, D.J.; Tran, L.M.; Li, Y.; Cai, H.; Morim, A.; Wang, S.; Plaisier, S.; Garraway, I.P.; Huang, J.; Graeber, T.G.; et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell 2011, 19, 792–804. [Google Scholar] [CrossRef] [Green Version]
- Marcelli, M.; Haidacher, S.J.; Plymate, S.R.; Birnbaum, R.S. Altered growth and insulin-like growth factor-binding protein-3 production in PC3 prostate carcinoma cells stably transfected with a constitutively active androgen receptor complementary deoxyribonucleic acid. Endocrinology 1995, 136, 1040–1048. [Google Scholar] [CrossRef]
- Xinzhou, H.; Ning, Y.; Ou, W.; Xiaodan, L.; Fumin, Y.; Huitu, L.; Wei, Z. RKIp inhibits the migration and invasion of human prostate cancer PC-3M cells through regulation of extracellular matrix. Mol. Biol. 2011, 45, 1004–1011. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, E.S.; Kang, H.E.; Kim, N.H.; Yook, J.I. Therapeutic implications of cancer epithelial-mesenchymal transition (EMT). Arch. Pharm Res. 2019, 42, 14–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, W.; Huan, M.; Yang, W.; Gao, Y.; Wang, K.; Xu, S.; Zhang, M.; Ma, J.; Wang, X.; Chen, Y.; et al. Snail promotes prostate cancer migration by facilitating SPOP ubiquitination and degradation. Biochem. Biophys. Res. Commun. 2020, 529, 799–804. [Google Scholar] [CrossRef]
- Beach, S.; Tang, H.; Park, S.; Dhillon, A.S.; Keller, E.T.; Kolch, W.; Yeung, K.C. Snail is a repressor of RKIP transcription in metastatic prostate cancer cells. Oncogene 2008, 27, 2243–2248. [Google Scholar] [CrossRef] [Green Version]
- Baritaki, S.; Yeung, K.; Palladino, M.; Berenson, J.; Bonavida, B. Pivotal roles of snail inhibition and RKIP induction by the proteasome inhibitor NPI-0052 in tumor cell chemoimmunosensitization. Cancer Res. 2009, 69, 8376–8385. [Google Scholar] [CrossRef] [Green Version]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Urrutia, E.; Bustamante Montes, L.P.; Ladron de Guevara Cervantes, D.; Perez-Plasencia, C.; Campos-Parra, A.D. Crosstalk Between Long Non-coding RNAs, Micro-RNAs and mRNAs: Deciphering Molecular Mechanisms of Master Regulators in Cancer. Front. Oncol. 2019, 9, 669. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Liu, X.H.; Zhu, H.C.; Wang, L.; Ning, J.Z.; Xiao, C.C. MiR-543 Promotes Proliferation and Epithelial-Mesenchymal Transition in Prostate Cancer via Targeting RKIP. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2017, 41, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Weng, X.D.; Wang, L.; Liu, X.H.; Zhu, H.C.; Guo, J.; Ning, J.Z.; Xiao, C.C. LncRNA XIST acts as a tumor suppressor in prostate cancer through sponging miR-23a to modulate RKIP expression. Oncotarget 2017, 8, 94358–94370. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, M.; Lai, T.H.; Zada, S.; Hwang, J.S.; Pham, T.M.; Yun, M.; Kim, D.R. Functional Linkage of RKIP to the Epithelial to Mesenchymal Transition and Autophagy during the Development of Prostate Cancer. Cancers 2018, 10, 273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aakula, A.; Kohonen, P.; Leivonen, S.K.; Makela, R.; Hintsanen, P.; Mpindi, J.P.; Martens-Uzunova, E.; Aittokallio, T.; Jenster, G.; Perala, M.; et al. Systematic Identification of MicroRNAs That Impact on Proliferation of Prostate Cancer Cells and Display Changed Expression in Tumor Tissue. Eur. Urol. 2016, 69, 1120–1128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Cai, L.; Tian, X.; Li, W. MiR-543 promotes tumorigenesis and angiogenesis in non-small cell lung cancer via modulating metastasis associated protein 1. Mol. Med. 2020, 26, 44. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Jiang, W.; Zhang, X.; Lu, Z.; Geng, Q.; Wang, W.; Li, N.; Cai, X. LINC-PINT alleviates lung cancer progression via sponging miR-543 and inducing PTEN. Cancer Med. 2020, 9, 1999–2009. [Google Scholar] [CrossRef] [Green Version]
- Su, D.W.; Li, X.; Chen, J.; Dou, J.; Fang, G.E.; Luo, C.J. MiR-543 inhibits proliferation and metastasis of human colorectal cancer cells by targeting PLAS3. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 8812–8821. [Google Scholar] [CrossRef]
- Liu, X.; Gan, L.; Zhang, J. miR-543 inhibites cervical cancer growth and metastasis by targeting TRPM7. Chem. Biol. Interact. 2019, 302, 83–92. [Google Scholar] [CrossRef]
- Fu, Q.; Liu, X.; Liu, Y.; Yang, J.; Lv, G.; Dong, S. MicroRNA-335 and -543 suppress bone metastasis in prostate cancer via targeting endothelial nitric oxide synthase. Int. J. Mol. Med. 2015, 36, 1417–1425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.C.; He, W.Y.; Dong, C.H.; Pei, L.; Ma, Y.L. lncRNA HCG11 regulates cell progression by targeting miR-543 and regulating AKT/mTOR pathway in prostate cancer. Cell Biol. Int. 2019, 43, 1453–1462. [Google Scholar] [CrossRef]
- Wang, X.; Yang, J.Y.; Cai, J.; Zhang, D.J.; Zhao, L.; Luo, L.H.; Xiong, Y.; Zhang, T.; Jin, M. MiR-543/Numb promotes proliferation, metastasis, and stem-like cell traits of prostate cancer cells. Am. J. Transl. Res. 2021, 13, 617–631. [Google Scholar] [PubMed]
- Baritaki, S.; Huerta-Yepez, S.; Sahakyan, A.; Karagiannides, I.; Bakirtzi, K.; Jazirehi, A.; Bonavida, B. Mechanisms of nitric oxide-mediated inhibition of EMT in cancer: Inhibition of the metastasis-inducer Snail and induction of the metastasis-suppressor RKIP. Cell Cycle 2010, 9, 4931–4940. [Google Scholar] [CrossRef] [Green Version]
- Della Pietra, E.; Simonella, F.; Bonavida, B.; Xodo, L.E.; Rapozzi, V. Repeated sub-optimal photodynamic treatments with pheophorbide a induce an epithelial mesenchymal transition in prostate cancer cells via nitric oxide. Nitric Oxide 2015, 45, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Rapozzi, V.; Varchi, G.; Della Pietra, E.; Ferroni, C.; Xodo, L.E. A photodynamic bifunctional conjugate for prostate cancer: An in vitro mechanistic study. Investig. New Drugs 2017, 35, 115–123. [Google Scholar] [CrossRef]
- Kashyap, V.; Bonavida, B. Role of YY1 in the pathogenesis of prostate cancer and correlation with bioinformatic data sets of gene expression. Genes Cancer 2014, 5, 71–83. [Google Scholar] [CrossRef] [Green Version]
- D’Este, F.; Della Pietra, E.; Badillo Pazmay, G.V.; Xodo, L.E.; Rapozzi, V. Role of nitric oxide in the response to photooxidative stress in prostate cancer cells. Biochem. Pharmacol. 2020, 182, 114205. [Google Scholar] [CrossRef]
- Chatterjee, D.; Bai, Y.; Wang, Z.; Beach, S.; Mott, S.; Roy, R.; Braastad, C.; Sun, Y.; Mukhopadhyay, A.; Aggarwal, B.B.; et al. RKIP sensitizes prostate and breast cancer cells to drug-induced apoptosis. J. Biol. Chem. 2004, 279, 17515–17523. [Google Scholar] [CrossRef] [Green Version]
- Woods Ignatoski, K.M.; Grewal, N.K.; Markwart, S.M.; Vellaichamy, A.; Chinnaiyan, A.M.; Yeung, K.; Ray, M.E.; Keller, E.T. Loss of Raf kinase inhibitory protein induces radioresistance in prostate cancer. Int. J. Radiat Oncol. Biol. Phys. 2008, 72, 153–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tannock, I.F.; de Wit, R.; Berry, W.R.; Horti, J.; Pluzanska, A.; Chi, K.N.; Oudard, S.; Theodore, C.; James, N.D.; Turesson, I.; et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N. Engl. J. Med. 2004, 351, 1502–1512. [Google Scholar] [CrossRef] [Green Version]
- Berthold, D.R.; Pond, G.R.; Soban, F.; de Wit, R.; Eisenberger, M.; Tannock, I.F. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer: Updated survival in the TAX 327 study. J. Clin. Oncol. 2008, 26, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.X.; Li, W.Z.; Guo, Y.L.; Chen, L.; Li, G.H.; Yu, J.J.; Shu, B.; Peng, S. Tumor suppressor RKIP inhibits prostate cancer cell metastasis and sensitizes prostate cancer cells to docetaxel treatment. Neoplasma 2018, 65, 228–233. [Google Scholar] [CrossRef]
- Caohuy, H.; Srivastava, M.; Pollard, H.B. Membrane fusion protein synexin (annexin VII) as a Ca2+/GTP sensor in exocytotic secretion. Proc. Natl. Acad. Sci. USA 1996, 93, 10797–10802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerke, V.; Moss, S.E. Annexins: From structure to function. Physiol. Rev. 2002, 82, 331–371. [Google Scholar] [CrossRef]
- Guo, C.; Liu, S.; Greenaway, F.; Sun, M.Z. Potential role of annexin A7 in cancers. Clin. Chim. Acta Int. J. Clin. Chem. 2013, 423, 83–89. [Google Scholar] [CrossRef]
- Xin, W.; Rhodes, D.R.; Ingold, C.; Chinnaiyan, A.M.; Rubin, M.A. Dysregulation of the annexin family protein family is associated with prostate cancer progression. Am. J. Pathol. 2003, 162, 255–261. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, M.; Bubendorf, L.; Srikantan, V.; Fossom, L.; Nolan, L.; Glasman, M.; Leighton, X.; Fehrle, W.; Pittaluga, S.; Raffeld, M.; et al. ANX7, a candidate tumor suppressor gene for prostate cancer. Proc. Natl. Acad. Sci. USA 2001, 98, 4575–4580. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, M.; Montagna, C.; Leighton, X.; Glasman, M.; Naga, S.; Eidelman, O.; Ried, T.; Pollard, H.B. Haploinsufficiency of Anx7 tumor suppressor gene and consequent genomic instability promotes tumorigenesis in the Anx7(+/−) mouse. Proc. Natl. Acad. Sci. USA 2003, 100, 14287–14292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torosyan, Y.; Simakova, O.; Naga, S.; Mezhevaya, K.; Leighton, X.; Diaz, J.; Huang, W.; Pollard, H.; Srivastava, M. Annexin-A7 protects normal prostate cells and induces distinct patterns of RB-associated cytotoxicity in androgen-sensitive and -resistant prostate cancer cells. Int. J. Cancer 2009, 125, 2528–2539. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, X.; Lin, Z.; Su, L.; Yan, S.; Zhao, B.; Miao, J. SEC-induced activation of ANXA7 GTPase suppresses prostate cancer metastasis. Cancer Lett. 2018, 416, 11–23. [Google Scholar] [CrossRef]
- Simon, J.A.; Lange, C.A. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat. Res. 2008, 647, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, R.; Zhang, Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 2004, 14, 155–164. [Google Scholar] [CrossRef]
- Pasini, D.; Di Croce, L. Emerging roles for Polycomb proteins in cancer. Curr. Opin. Genet. Dev. 2016, 36, 50–58. [Google Scholar] [CrossRef]
- Yamagishi, M.; Uchimaru, K. Targeting EZH2 in cancer therapy. Curr. Opin. Oncol. 2017, 29, 375–381. [Google Scholar] [CrossRef]
- Kim, K.H.; Roberts, C.W. Targeting EZH2 in cancer. Nat. Med. 2016, 22, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Liu, C.; Zhou, B.; Bi, L.; Huang, H.; Lin, T.; Xu, K. Role of EZH2 in the growth of prostate cancer stem cells isolated from LNCaP cells. Int. J. Mol. Sci. 2013, 14, 11981–11993. [Google Scholar] [CrossRef] [Green Version]
- Ren, G.; Baritaki, S.; Marathe, H.; Feng, J.; Park, S.; Beach, S.; Bazeley, P.S.; Beshir, A.B.; Fenteany, G.; Mehra, R.; et al. Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer Res. 2012, 72, 3091–3104. [Google Scholar] [CrossRef] [Green Version]
- Angell, H.; Galon, J. From the immune contexture to the Immunoscore: The role of prognostic and predictive immune markers in cancer. Curr. Opin. Immunol. 2013, 25, 261–267. [Google Scholar] [CrossRef]
- Gentles, A.J.; Newman, A.M.; Liu, C.L.; Bratman, S.V.; Feng, W.; Kim, D.; Nair, V.S.; Xu, Y.; Khuong, A.; Hoang, C.D.; et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015, 21, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Ruiz de Porras, V.; Pardo, J.C.; Notario, L.; Etxaniz, O.; Font, A. Immune Checkpoint Inhibitors: A Promising Treatment Option for Metastatic Castration-Resistant Prostate Cancer? Int. J. Mol. Sci. 2021, 22, 4712. [Google Scholar] [CrossRef] [PubMed]
- Ru, B.; Wong, C.N.; Tong, Y.; Zhong, J.Y.; Zhong, S.S.W.; Wu, W.C.; Chu, K.C.; Wong, C.Y.; Lau, C.Y.; Chen, I.; et al. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics 2019, 35, 4200–4202. [Google Scholar] [CrossRef] [PubMed]
- Dyck, L.; Mills, K.H.G. Immune checkpoints and their inhibition in cancer and infectious diseases. Eur. J. Immunol. 2017, 47, 765–779. [Google Scholar] [CrossRef]
- Rowshanravan, B.; Halliday, N.; Sansom, D.M. CTLA-4: A moving target in immunotherapy. Blood 2018, 131, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Dougall, W.C.; Kurtulus, S.; Smyth, M.J.; Anderson, A.C. TIGIT and CD96: New checkpoint receptor targets for cancer immunotherapy. Immunol. Rev. 2017, 276, 112–120. [Google Scholar] [CrossRef]
- Cannarile, M.A.; Weisser, M.; Jacob, W.; Jegg, A.M.; Ries, C.H.; Ruttinger, D. Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J. Immunother Cancer 2017, 5, 53. [Google Scholar] [CrossRef]
- Abida, W.; Cyrta, J.; Heller, G.; Prandi, D.; Armenia, J.; Coleman, I.; Cieslik, M.; Benelli, M.; Robinson, D.; Van Allen, E.M.; et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 11428–11436. [Google Scholar] [CrossRef] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wing, K.; Onishi, Y.; Prieto-Martin, P.; Yamaguchi, T.; Miyara, M.; Fehervari, Z.; Nomura, T.; Sakaguchi, S. CTLA-4 control over Foxp3+ regulatory T cell function. Science 2008, 322, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Keller, E.T.; Fu, Z.; Brennan, M. The role of Raf kinase inhibitor protein (RKIP) in health and disease. Biochem. Pharmacol. 2004, 68, 1049–1053. [Google Scholar] [CrossRef]
- Ku, S.Y.; Rosario, S.; Wang, Y.; Mu, P.; Seshadri, M.; Goodrich, Z.W.; Goodrich, M.M.; Labbe, D.P.; Gomez, E.C.; Wang, J.; et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017, 355, 78–83. [Google Scholar] [CrossRef] [Green Version]
- Mostaghel, E.A.; Page, S.T.; Lin, D.W.; Fazli, L.; Coleman, I.M.; True, L.D.; Knudsen, B.; Hess, D.L.; Nelson, C.C.; Matsumoto, A.M.; et al. Intraprostatic androgens and androgen-regulated gene expression persist after testosterone suppression: Therapeutic implications for castration-resistant prostate cancer. Cancer Res. 2007, 67, 5033–5041. [Google Scholar] [CrossRef] [Green Version]
- Montgomery, R.B.; Mostaghel, E.A.; Vessella, R.; Hess, D.L.; Kalhorn, T.F.; Higano, C.S.; True, L.D.; Nelson, P.S. Maintenance of intratumoral androgens in metastatic prostate cancer: A mechanism for castration-resistant tumor growth. Cancer Res. 2008, 68, 4447–4454. [Google Scholar] [CrossRef] [Green Version]
- Granovsky, A.E.; Rosner, M.R. Raf kinase inhibitory protein: A signal transduction modulator and metastasis suppressor. Cell Res. 2008, 18, 452–457. [Google Scholar] [CrossRef] [Green Version]
- Granovsky, A.E.; Clark, M.C.; McElheny, D.; Heil, G.; Hong, J.; Liu, X.; Kim, Y.; Joachimiak, G.; Joachimiak, A.; Koide, S.; et al. Raf kinase inhibitory protein function is regulated via a flexible pocket and novel phosphorylation-dependent mechanism. Mol. Cell. Biol. 2009, 29, 1306–1320. [Google Scholar] [CrossRef] [Green Version]
- Skinner, J.J.; Rosner, M.R. RKIP structure drives its function: A three-state model for regulation of RKIP. Crit. Rev. Oncog. 2014, 19, 483–488. [Google Scholar] [CrossRef] [Green Version]
- Pnueli, L.; Carmel-Goren, L.; Hareven, D.; Gutfinger, T.; Alvarez, J.; Ganal, M.; Zamir, D.; Lifschitz, E. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 1998, 125, 1979–1989. [Google Scholar] [CrossRef] [PubMed]



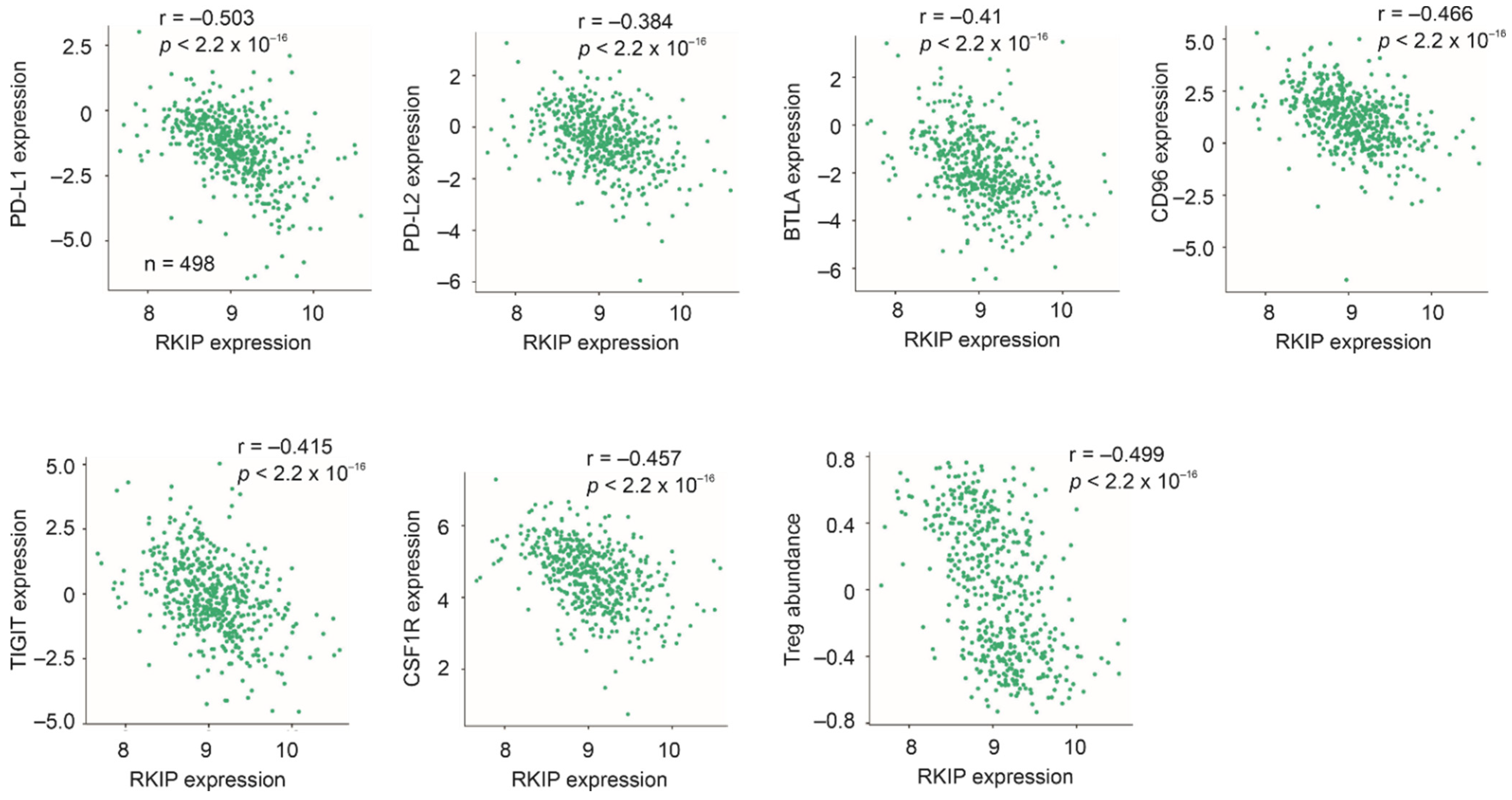
| Cancer Type | Population (n) | Spearman r | p Value |
|---|---|---|---|
| Bladder uroghelial carcinoma | 408 | −0.432 b; −0.469 c | <2.2 × 10−16; <2.2 × 10−16 |
| Breast cabcer | 1100 | −0.41; −0.4 | <2.2 × 10−16; <2.2 × 10−16 |
| Lung adenocarcinoma | 517 | −0.428; −0.325 | <2.2 × 10−16; 4.47 × 10−14 |
| Rectum adenocarcinoma | 167 | −0.414; −0.36 | 3.66 × 10−8; 2.09 × 10−6 |
| Thyroid carcinoma | 509 | −0.55; −0.415 | <2.2 × 10−16; <2.2 × 10−16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, Y.; Lin, X.; Kapoor, A.; Gu, Y.; Xu, H.; Major, P.; Tang, D. Insights of RKIP-Derived Suppression of Prostate Cancer. Cancers 2021, 13, 6388. https://doi.org/10.3390/cancers13246388
Dong Y, Lin X, Kapoor A, Gu Y, Xu H, Major P, Tang D. Insights of RKIP-Derived Suppression of Prostate Cancer. Cancers. 2021; 13(24):6388. https://doi.org/10.3390/cancers13246388
Chicago/Turabian StyleDong, Ying, Xiaozeng Lin, Anil Kapoor, Yan Gu, Hui Xu, Pierre Major, and Damu Tang. 2021. "Insights of RKIP-Derived Suppression of Prostate Cancer" Cancers 13, no. 24: 6388. https://doi.org/10.3390/cancers13246388







