Sequential Isolation and Characterization of Single CTCs and Large CTC Clusters in Metastatic Colorectal Cancer Patients
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Blood Samples Collection
2.2. Establishment of a Customized Filtration Method for the Isolation of CTCs Clusters
2.3. Sequential Isolation of Single CTCs and of CTC Clusters
2.4. Immunofluorescence Staining
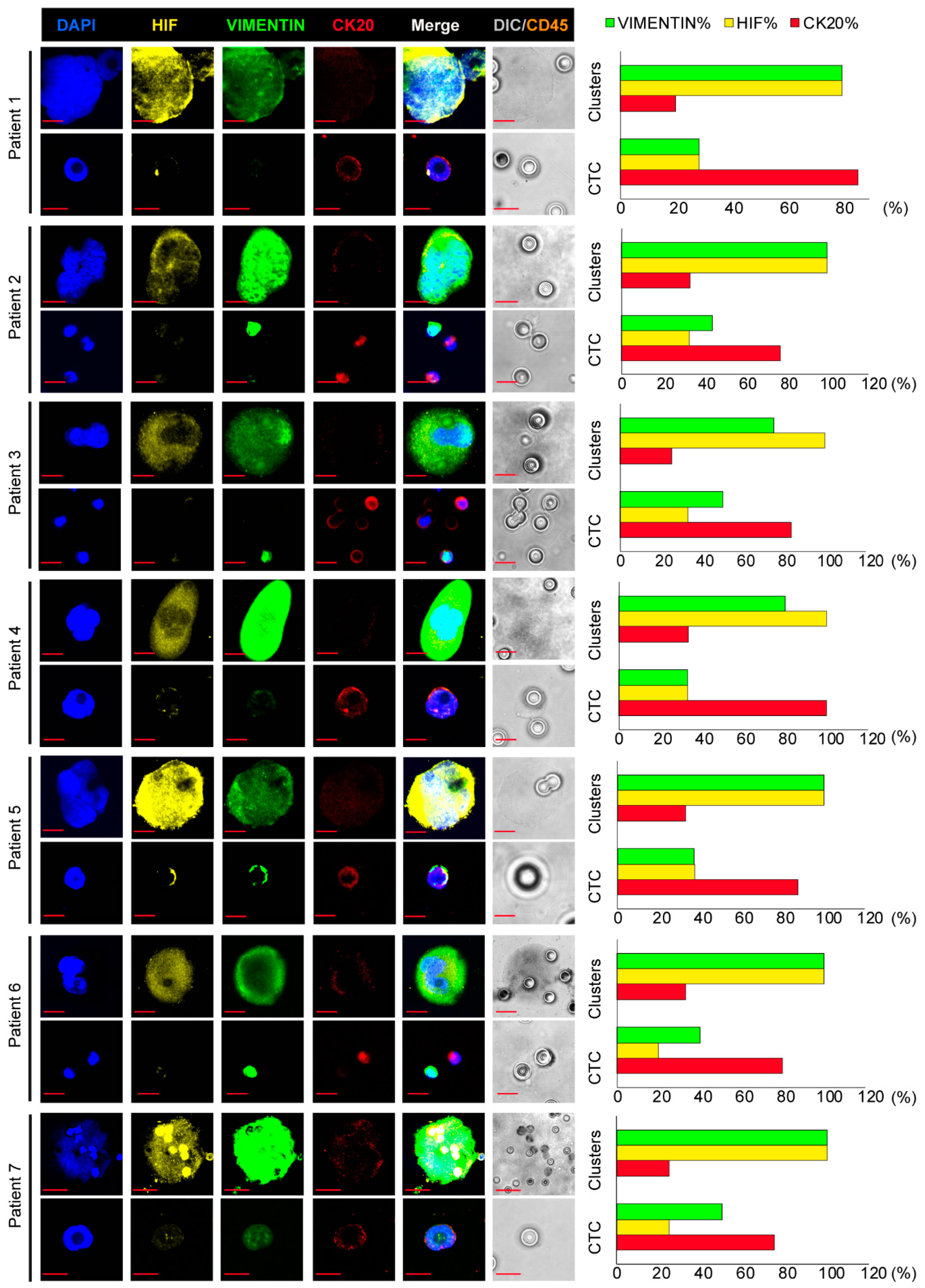
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paoletti, C.; Hayes, D.F. Circulating Tumor Cells. In Novel Biomarkers in the Continuum of Breast Cancer; Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2015; Volume 882, pp. 235–258. [Google Scholar] [CrossRef]
- Castro-Giner, F.; Aceto, N. Tracking cancer progression: From circulating tumor cells to metastasis. Genome Med. 2020, 12, 31. [Google Scholar] [CrossRef] [Green Version]
- Massagué, J.; Obenauf, A.C. Metastatic colonization by circulating tumour cells. Nature 2016, 529, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-C.; Zhang, X.-F.; Peng, J.; Li, X.-F.; Wang, A.-L.; Bie, Y.-Q.; Shi, L.-H.; Lin, M.-B. Survival Mechanisms and Influence Factors of Circulating Tumor Cells. BioMed Res. Int. 2018, 2018, 6304701. [Google Scholar] [CrossRef] [PubMed]
- Strilic, B.; Offermanns, S. Intravascular Survival and Extravasation of Tumor Cells. Cancer Cell 2017, 32, 282–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aceto, N.; Bardia, A.; Miyamoto, D.T.; Donaldson, M.C.; Wittner, B.S.; Spencer, J.A.; Yu, M.; Pely, A.; Engstrom, A.; Zhu, H.; et al. Circulating Tumor Cell Clusters Are Oligoclonal Precursors of Breast Cancer Metastasis. Cell 2014, 158, 1110–1122. [Google Scholar] [CrossRef] [Green Version]
- Amintas, S.; Bedel, A.; Moreau-Gaudry, F.; Boutin, J.; Buscail, L.; Merlio, J.-P.; Vendrely, V.; Dabernat, S.; Buscail, E. Circulating Tumor Cell Clusters: United We Stand Divided We Fall. Int. J. Mol. Sci. 2020, 21, 2653. [Google Scholar] [CrossRef] [Green Version]
- Kapeleris, J.; Zou, H.; Qi, Y.; Gu, Y.; Li, J.; Schoning, J.; Monteiro, M.J.; Gu, W. Cancer stemness contributes to cluster formation of colon cancer cells and high metastatic potentials. Clin. Exp. Pharmacol. Physiol. 2019, 47, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Zeng, H.; Gu, X.; Ma, W. Circulating tumor cell clusters-associated gene plakoglobin and breast cancer survival. Breast Cancer Res. Treat. 2015, 151, 491–500. [Google Scholar] [CrossRef]
- Chang, P.-H.; Chen, M.-C.; Tsai, Y.-P.; Tan, G.Y.T.; Hsu, P.-H.; Jeng, Y.-M.; Tsai, Y.-F.; Yang, M.-H.; Hwang-Verslues, W.W. Interplay between desmoglein2 and hypoxia controls metastasis in breast cancer. Proc. Natl. Acad. Sci. USA 2021, 118, e2014408118. [Google Scholar] [CrossRef]
- Dementeva, N.; Kokova, D.; Mayboroda, O. Current Methods of the Circulating Tumor Cells (CTC) Analysis: A Brief Overview. Curr. Pharm. Des. 2017, 23, 4726–4728. [Google Scholar] [CrossRef]
- Ferreira, M.M.; Ramani, V.C.; Jeffrey, S.S. Circulating tumor cell technologies. Mol. Oncol. 2016, 10, 374–394. [Google Scholar] [CrossRef] [Green Version]
- Hendricks, A.; Brandt, B.; Geisen, R.; Dall, K.; Röder, C.; Schafmayer, C.; Becker, T.; Hinz, S.; Sebens, S. Isolation and Enumeration of CTC in Colorectal Cancer Patients: Introduction of a Novel Cell Imaging Approach and Comparison to Cellular and Molecular Detection Techniques. Cancers 2020, 12, 2643. [Google Scholar] [CrossRef]
- DeSitter, I.; Guerrouahen, B.S.; Benali-Furet, N.; Wechsler, J.; Jänne, P.A.; Kuang, Y.; Yanagita, M.; Wang, L.; Berkowitz, J.A.; Distel, R.J.; et al. A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer. Res. 2011, 31, 427–441. [Google Scholar]
- Hong, Y.; Fang, F.; Zhang, Q. Circulating tumor cell clusters: What we know and what we expect (Review). Int. J. Oncol. 2016, 49, 2206–2216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankó, P.; Lee, S.Y.; Nagygyörgy, V.; Zrínyi, M.; Chae, C.H.; Cho, D.H.; Telekes, A. Technologies for circulating tumor cell separation from whole blood. J. Hematol. Oncol. 2019, 12, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rushton, A.; Nteliopoulos, G.; Shaw, J.; Coombes, R. A Review of Circulating Tumour Cell Enrichment Technologies. Cancers 2021, 13, 970. [Google Scholar] [CrossRef]
- Raimondi, C.; Carpino, G.; Nicolazzo, C.; Gradilone, A.; Gianni, W.; Gelibter, A.; Gaudio, E.; Cortesi, E.; Gazzaniga, P. PD-L1 and epithelial-mesenchymal transition in circulating tumor cells from non-small cell lung cancer patients: A molecular shield to evade immune system? Oncoimmunology 2017, 6, e1315488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolazzo, C.; Raimondi, C.; Gradilone, A.; Emiliani, A.; Zeuner, A.; Francescangeli, F.; Belardinilli, F.; Seminara, P.; Loreni, F.; Magri, V.; et al. Circulating Tumor Cells in Right- and Left-Sided Colorectal Cancer. Cancers 2019, 11, 1042. [Google Scholar] [CrossRef] [Green Version]
- Gazzaniga, P.; Raimondi, C.; Nicolazzo, C.; Carletti, R.; Di Gioia, C.; Gradilone, A.; Cortesi, E. The rationale for liquid biopsy in colorectal cancer: A focus on circulating tumor cells. Expert Rev. Mol. Diagn. 2015, 15, 925–932. [Google Scholar] [CrossRef] [Green Version]
- Divella, R.; Daniele, A.; Abbate, I.; Bellizzi, A.; Savino, E.; Simone, G.; Giannone, G.; Giuliani, F.; Fazio, V.; Gadaleta-Caldarola, G.; et al. The presence of clustered circulating tumor cells (CTCs) and circulating cytokines define an aggressive phenotype in metastatic colorectal cancer. Cancer Causes Control 2014, 25, 1531–1541. [Google Scholar] [CrossRef]
- Donato, C.; Kunz, L.; Castro-Giner, F.; Paasinen-Sohns, A.; Strittmatter, K.; Szczerba, B.M.; Scherrer, R.; Di Maggio, N.; Heusermann, W.; Biehlmaier, O.; et al. Hypoxia Triggers the Intravasation of Clustered Circulating Tumor Cells. Cell Rep. 2020, 32, 108105. [Google Scholar] [CrossRef] [PubMed]
- Petrova, V.; Annicchiarico-Petruzzelli, M.; Melino, G.; Amelio, I. The hypoxic tumour microenvironment. Oncogenesis 2018, 7, 10. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | No. (%) |
|---|---|
| Age (in years) | |
| Mean | 67.7 |
| Range | 55–84 |
| PS | |
| 0 | 4 (57) |
| 1 | 3 (43) |
| Sex | |
| Male | 3 (43) |
| Female | 4 (57) |
| Colorectal cancer stage | |
| IV | 7 (100) |
| Right-sided | 4 (57) |
| Left-sided | 3 (43) |
| Mutations | |
| RAS | 4 (57) |
| BRAF | 1 (14) |
| Patient | CTC | CTC Cluster | ||||||
|---|---|---|---|---|---|---|---|---|
| NT | CK20 (N) | HIF-1α (N) | VIM (N) | NT | CK20 (N) | HIF-1α (N) | VIM (N) | |
| 14AA6844 | 7 | 6 | 2 | 2 | 5 | 1 | 4 | 4 |
| 14AA6865 | 9 | 7 | 3 | 4 | 3 | 1 | 3 | 3 |
| 14AA6922 | 6 | 5 | 2 | 3 | 4 | 1 | 4 | 3 |
| 15AA0421 | 3 | 3 | 1 | 1 | 6 | 2 | 5 | 4 |
| 15AA0433 | 8 | 7 | 3 | 3 | 6 | 2 | 6 | 6 |
| 15AA0814 | 5 | 4 | 1 | 2 | 3 | 1 | 2 | 2 |
| 15AA0924 | 4 | 3 | 1 | 2 | 4 | 1 | 2 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Francescangeli, F.; Magri, V.; De Angelis, M.L.; De Renzi, G.; Gandini, O.; Zeuner, A.; Gazzaniga, P.; Nicolazzo, C. Sequential Isolation and Characterization of Single CTCs and Large CTC Clusters in Metastatic Colorectal Cancer Patients. Cancers 2021, 13, 6362. https://doi.org/10.3390/cancers13246362
Francescangeli F, Magri V, De Angelis ML, De Renzi G, Gandini O, Zeuner A, Gazzaniga P, Nicolazzo C. Sequential Isolation and Characterization of Single CTCs and Large CTC Clusters in Metastatic Colorectal Cancer Patients. Cancers. 2021; 13(24):6362. https://doi.org/10.3390/cancers13246362
Chicago/Turabian StyleFrancescangeli, Federica, Valentina Magri, Maria Laura De Angelis, Gianluigi De Renzi, Orietta Gandini, Ann Zeuner, Paola Gazzaniga, and Chiara Nicolazzo. 2021. "Sequential Isolation and Characterization of Single CTCs and Large CTC Clusters in Metastatic Colorectal Cancer Patients" Cancers 13, no. 24: 6362. https://doi.org/10.3390/cancers13246362






