The Modes of Dysregulation of the Proto-Oncogene T-Cell Leukemia/Lymphoma 1A
Abstract
:Simple Summary
Abstract
1. Introduction
2. The Normal and Tumor-Associated Expression of TCL1A
2.1. The Physiological Expression of TCL1A
2.2. Expression of TCL1A in Hematologic Malignancies
2.3. Expression of TCL1A in Solid Tumors
2.4. Clinical Impact of Detection of TCL1A
3. The Physiological and Disease-Associated Function of TCL1A
3.1. The Functional Role of TCL1A in Embryonic Development and Stemness
3.2. The Functional Role of TCL1A in Cancer Signaling and Pathogenesis
4. The Modes of TCL1A (Dys)Regulation
4.1. TCL1A Transcriptional Regulation in Embryonic Stem Cells and Cancer Stem Cells
4.2. TCL1A Promoter Activation
4.3. Posttranscriptional Regulation of TCL1A by Micro RNAs
4.4. Posttranslational Regulation of TCL1A
5. Exogeneous Triggers of TCL1A-Regulating Mechanisms
5.1. Regulation of TCL1A Levels by the Microenvironment
5.2. Alterations of TCL1A Expression in EBV Infection
6. Discussion
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Virgilio, L.; Narducci, M.G.; Isobe, M.; Billips, L.G.; Cooper, M.D.; Croce, C.M.; Russo, G. Identification of the TCL1 gene involved in T-cell malignancies. Proc. Natl. Acad. Sci. USA 1994, 91, 12530–12534. [Google Scholar] [CrossRef] [Green Version]
- Russo, G.; Isobe, M.; Gatti, R.; Finan, J.; Batuman, O.; Huebner, K.; Nowell, P.C.; Croce, C.M. Molecular analysis of a t(14;14) translocation in leukemic T-cells of an ataxia telangiectasia patient. Proc. Natl. Acad. Sci. USA 1989, 86, 602–606. [Google Scholar] [CrossRef] [Green Version]
- Virgilio, L.; Isobe, M.; Narducci, M.G.; Carotenuto, P.; Camerini, B.; Kurosawa, N.; Abbas-ar-Rushdi; Croce, C.M.; Russo, G. Chromosome walking on the TCL1 locus involved in T-cell neoplasia. Proc. Natl. Acad. Sci. USA 1993, 90, 9275–9279. [Google Scholar] [CrossRef] [Green Version]
- Teitell, M.A. The TCL1 family of oncoproteins: Co-activators of transformation. Nat. Rev. Cancer 2005, 5, 640–648. [Google Scholar] [CrossRef]
- Pekarsky, Y.; Hallas, C.; Isobe, M.; Russo, G.; Croce, C.M. Abnormalities at 14q32.1 in T cell malignancies involve two oncogenes. Proc. Natl. Acad. Sci. USA 1999, 96, 2949–2951. [Google Scholar] [CrossRef] [Green Version]
- Stern, M.H.; Soulier, J.; Rosenzwajg, M.; Nakahara, K.; Canki-Klain, N.; Aurias, A.; Sigaux, F.; Kirsch, I.R. MTCP-1: A novel gene on the human chromosome Xq28 translocated to the T cell receptor alpha/delta locus in mature T cell proliferations. Oncogene 1993, 8, 2475–2483. [Google Scholar]
- Fu, Z.Q.; Du Bois, G.C.; Song, S.P.; Kulikovskaya, I.; Virgilio, L.; Rothstein, J.L.; Croce, C.M.; Weber, I.T.; Harrison, R.W. Crystal structure of MTCP-1: Implications for role of TCL-1 and MTCP-1 in T cell malignancies. Proc. Natl. Acad. Sci. USA 1998, 95, 3413–3418. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.M.; Narducci, M.G.; Lazzeri, C.; Mongiovì, A.M.; Caprini, E.; Bresin, A.; Martelli, F.; Rothstein, J.; Croce, C.M.; Cooper, M.D.; et al. Impaired T- and B-cell development in Tcl1-deficient mice. Blood 2005, 105, 1288–1294. [Google Scholar] [CrossRef] [Green Version]
- Narducci, M.G.; Fiorenza, M.T.; Kang, S.M.; Bevilacqua, A.; Di Giacomo, M.; Remotti, D.; Picchio, M.C.; Fidanza, V.; Cooper, M.D.; Croce, C.M.; et al. TCL1 participates in early embryonic development and is overexpressed in human seminomas. Proc. Natl. Acad. Sci. USA 2002, 99, 11712–11717. [Google Scholar] [CrossRef] [Green Version]
- Fink, S.R.; Paternoster, S.F.; Smoley, S.A.; Flynn, H.C.; Geyer, S.M.; Shanafelt, T.D.; Lee, Y.K.; Jelinek, D.F.; Kay, N.E.; Dewald, G.W. Fluorescent-labeled DNA probes applied to novel biological aspects of B-cell chronic lymphocytic leukemia. Leuk. Res. 2005, 29, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Pekarsky, Y.; Palamarchuk, A.; Maximov, V.; Efanov, A.; Nazaryan, N.; Santanam, U.; Rassenti, L.; Kipps, T.; Croce, C.M. Tcl1 functions as a transcriptional regulator and is directly involved in the pathogenesis of CLL. Proc. Natl. Acad. Sci. USA 2008, 105, 19643–19648. [Google Scholar] [CrossRef] [Green Version]
- Herling, M.; Patel, K.A.; Hsi, E.D.; Chang, K.C.; Rassidakis, G.Z.; Ford, R.; Jones, D. TCL1 in B-cell tumors retains its normal B-cell pattern of regulation and is a marker of differentiation stage. Am. J. Surg. Pathol. 2007, 31, 1123–1129. [Google Scholar] [CrossRef] [PubMed]
- Virgilio, L.; Lazzeri, C.; Bichi, R.; Nibu, K.I.; Narducci, M.G.; Russo, G.; Rothstein, J.L.; Croce, C.M. Deregulated expression of TCL1 causes T cell leukemia in mice. Proc. Natl. Acad. Sci. USA 1998, 95, 3885–3889. [Google Scholar] [CrossRef] [Green Version]
- Gritti, C.; Dastot, H.; Soulier, J.; Janin, A.; Daniel, M.T.; Madani, A.M.; Grimber, G.; Briand, P.; Sigaux, F.; Stern, M.H. Transgenic mice for MTCP1 develop T-cell prolymphocytic leukemia. Blood 1998, 92, 368–373. [Google Scholar] [CrossRef]
- Hoyer, K.K.; French, S.W.; Turner, D.E.; Nguyen, M.T.N.; Renard, M.; Malone, C.S.; Knoetig, S.; Qi, C.F.; Su, T.T.; Cheroutre, H.; et al. Dysregulated TCL1 promotes multiple classes of mature B cell lymphoma. Proc. Natl. Acad. Sci. USA 2002, 99, 14392–14397. [Google Scholar] [CrossRef] [Green Version]
- Bichi, R.; Shinton, S.A.; Martin, E.S.; Koval, A.; Calin, G.A.; Cesari, R.; Russo, G.; Hardy, R.R.; Croce, C.M. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc. Natl. Acad. Sci. USA 2002, 99, 6955–6960. [Google Scholar] [CrossRef] [Green Version]
- Simonetti, G.; Bertilaccio, M.T.S.; Ghia, P.; Klein, U. Mouse models in the study of chronic lymphocytic leukemia pathogenesis and therapy. Blood 2014, 124, 1010–1019. [Google Scholar] [CrossRef] [Green Version]
- Bresin, A.; D’Abundo, L.; Narducci, M.G.; Fiorenza, M.T.; Croce, C.M.; Negrini, M.; Russo, G. TCL1 transgenic mouse model as a tool for the study of therapeutic targets and microenvironment in human B-cell chronic lymphocytic leukemia. Cell Death Dis. 2016, 7, e2071. [Google Scholar] [CrossRef] [Green Version]
- Fedorchenko, O.; Stiefelhagen, M.; Peer-Zada, A.A.; Barthel, R.; Mayer, P.; Eckei, L.; Breuer, A.; Crispatzu, G.; Rosen, N.; Landwehr, T.; et al. CD44 regulates the apoptotic response and promotes disease development in chronic lymphocytic leukemia. Blood 2013, 121, 4126–4136. [Google Scholar] [CrossRef] [Green Version]
- Lewis, R.; Maurer, H.C.; Singh, N.; Gonzalez-Menendez, I.; Wirth, M.; Schick, M.; Zhang, L.; Isaakidis, K.; Scherger, A.K.; Schulze, V.; et al. CXCR4 hyperactivation cooperates with TCL1 in CLL development and aggressiveness. Leukemia 2021, 35, 2895–2905. [Google Scholar] [CrossRef]
- Holler, C.; Piñón, J.D.; Denk, U.; Heyder, C.; Hofbauer, S.; Greil, R.; Egle, A. PKC2 is essential for the development of chronic lymphocytic leukemia in the TCL1 transgenic mouse model: Validation of PKC2 as a therapeutic target in chronic lymphocytic leukemia. Blood 2009, 113, 2791–2794. [Google Scholar] [CrossRef] [Green Version]
- Kohlhaas, V.; Blakemore, S.J.; Al-Maarri, M.; Nickel, N.; Pal, M.; Roth, A.; Hövelmeyer, N.; Schäfer, S.C.; Knittel, G.; Lohneis, P.; et al. Active Akt signaling triggers CLL toward Richter transformation via overactivation of Notch1. Blood 2021, 137, 646–660. [Google Scholar] [CrossRef] [PubMed]
- Knittel, G.; Rehkämper, T.; Korovkina, D.; Liedgens, P.; Fritz, C.; Torgovnick, A.; Al-Baldawi, Y.; Al-Maarri, M.; Cun, Y.; Fedorchenko, O.; et al. Two mouse models reveal an actionable PARP1 dependence in aggressive chronic lymphocytic leukemia. Nat. Commun. 2017, 8, 153. [Google Scholar] [CrossRef]
- Herling, M.; Patel, K.A.; Weit, N.; Lilienthal, N.; Hallek, M.; Keating, M.J.; Jones, D. High TCL1 levels are a marker of B-cell receptor pathway responsiveness and adverse outcome in chronic lymphocytic leukemia. Blood 2009, 114, 4675–4686. [Google Scholar] [CrossRef]
- Ravandi, F.; O’Brien, S.; Jones, D.; Lerner, S.; Faderl, S.; Ferrajoli, A.; Wierda, W.; Garcia-Manero, G.; Thomas, D.; Koller, C.; et al. T-cell prolymphocytic leukemia: A single-institution experience. Clin. Lymphoma Myeloma 2005, 6, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Herling, M.; Patel, K.A.; Teitell, M.A.; Konopleva, M.; Ravandi, F.; Kobayashi, R.; Jones, D. High TCL1 expression and intact T-cell receptor signaling define a hyperproliferative subset of T-cell prolymphocytic leukemia. Blood 2008, 111, 328–337. [Google Scholar] [CrossRef]
- Schrader, A.; Crispatzu, G.; Oberbeck, S.; Mayer, P.; Pützer, S.; Von Jan, J.; Vasyutina, E.; Warner, K.; Weit, N.; Pflug, N.; et al. Actionable perturbations of damage responses by TCL1/ATM and epigenetic lesions form the basis of T-PLL. Nat. Commun. 2018, 9, 697. [Google Scholar] [CrossRef]
- Herling, M.; Patel, K.A.; Khalili, J.; Schlette, E.; Kobayashi, R.; Medeiros, L.J.; Jones, D. TCL1 shows a regulated expression pattern in chronic lymphocytic leukemia that correlates with molecular subtypes and proliferative state. Leukemia 2006, 20, 280–285. [Google Scholar] [CrossRef] [Green Version]
- Vasyutina, E.; Boucas, J.M.; Bloehdorn, J.; Aszyk, C.; Crispatzu, G.; Stiefelhagen, M.; Breuer, A.; Mayer, P.; Lengerke, C.; Döhner, H.; et al. The regulatory interaction of EVI1 with the TCL1A oncogene impacts cell survival and clinical outcome in CLL. Leukemia 2015, 29, 2003–2014. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yan, X.; Liu, L.; Huang, L.; Yin, M.; Pan, C.; Zhang, P.; Qin, H. T-cell leukemia/lymphoma-1A predicts the clinical outcome for patients with stage II/III colorectal cancer. Biomed. Pharmacother. 2017, 88, 924–930. [Google Scholar] [CrossRef]
- Narducci, M.G.; Virgilio, L.; Engiles, J.B.; Buchberg, A.M.; Billips, L.; Facchiano, A.; Croce, C.M.; Russo, G.; Rothstein, J.L. The murine Tcl1 oncogene: Embryonic and lymphoid cell expression. Oncogene 1997, 15, 919–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herling, M.; Teitell, M.A.; Shen, R.R.; Medeiros, L.J.; Jones, D. TCL1 expression in plasmacytoid dendritic cells (DC2s) and the related CD4+ CD56+ blastic tumors of skin. Blood 2003, 101, 5007–5009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoyer, K.K.; Herling, M.; Bagrintseva, K.; Dawson, D.W.; French, S.W.; Renard, M.; Weinger, J.G.; Jones, D.; Teitell, M.A. T Cell Leukemia-1 Modulates TCR Signal Strength and IFN-γ Levels through Phosphatidylinositol 3-Kinase and Protein Kinase C Pathway Activation. J. Immunol. 2005, 175, 864–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, P.; Cieslak, A.; Bernhart, S.H.; Toprak, U.H.; Wagener, R.; López, C.; Wiehle, L.; Bens, S.; Altmüller, J.; Franitza, M.; et al. Reconstruction of rearranged T-cell receptor loci by whole genome and transcriptome sequencing gives insights into the initial steps of T-cell prolymphocytic leukemia. Genes Chromosomes Cancer 2020, 59, 261–267. [Google Scholar] [CrossRef]
- Said, J.W.; Hoyer, K.K.; French, S.W.; Rosenfelt, L.; Garcia-Lloret, M.; Koh, P.J.; Cheng, T.C.; Sulur, G.G.; Pinkus, G.S.; Kuehl, W.M.; et al. TCL 1 oncogene expression in B cell subsets from lymphoid hyperplasia and distinct classes of B cell lymphoma. Lab. Investig. 2001, 81, 555–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saliba, J.; Saint-Martin, C.; Di Stefano, A.; Lenglet, G.; Marty, C.; Keren, B.; Pasquier, F.; Della Valle, V.; Secardin, L.; Leroy, G.; et al. Germline duplication of ATG2B and GSKIP predisposes to familial myeloid malignancies. Nat. Genet. 2015, 47, 1131–1140. [Google Scholar] [CrossRef]
- Babushok, D.V.; Stanley, N.L.; Morrissette, J.J.D.; Lieberman, D.B.; Olson, T.S.; Chou, S.T.; Hexner, E.O. Germline duplication of ATG2B and GSKIP genes is not required for the familial myeloid malignancy syndrome associated with the duplication of chromosome 14q32. Leukemia 2018, 32, 2720–2723. [Google Scholar] [CrossRef]
- Aggarwal, M.; Villuendas, R.; Gomez, G.; Rodriguez-Pinilla, S.M.; Sanchez-Beato, M.; Alvarez, D.; Martinez, N.; Rodriguez, A.; Castillo, M.E.; Camacho, F.I.; et al. TCL1A expression delineates biological and clinical variability in B-cell lymphoma. Mod. Pathol. 2009, 22, 206–215. [Google Scholar] [CrossRef] [Green Version]
- Lau, S.K.; Weiss, L.M.; Chu, P.G. TCL1 protein expression in testicular germ cell tumors. Am. J. Clin. Pathol. 2010, 133, 762–766. [Google Scholar] [CrossRef]
- Cao, D.; Lane, Z.; Allan, R.W.; Wang, P.; Guo, C.C.; Peng, Y.; Li, J. TCL1 is a diagnostic marker for intratubular germ cell neoplasia and classic seminoma. Histopathology 2010, 57, 152–157. [Google Scholar] [CrossRef]
- Lock, R.B. TCL1: A new drug target in lymphoid and germ-cell malignancies? Int. J. Biochem. Cell Biol. 2003, 35, 1614–1618. [Google Scholar] [CrossRef]
- Amini, S.; Fathi, F.; Mobalegi, J.; Sofimajidpour, H.; Ghadimi, T. The expressions of stem cell markers: Oct4, Nanog, Sox2, nucleostemin, Bmi, Zfx, Tcl1, Tbx3, Dppa4, and Esrrb in bladder, colon, and prostate cancer, and certain cancer cell lines. Anat. Cell Biol. 2014, 47, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.Q.; Ongkeko, W.M.; Chen, L.; Yang, Z.F.; Lu, P.; Chen, K.K.; Lopez, J.P.; Poon, R.T.P.; Fan, S.T. Octamer 4 (Oct4) mediates chemotherapeutic drug resistance in liver cancer cells through a potential Oct4-AKT-ATP-binding cassette G2 pathway. Hepatology 2010, 52, 528–539. [Google Scholar] [CrossRef]
- Hong, X.; Song, R.; Song, H.; Zheng, T.; Wang, J.; Liang, Y.; Qi, S.; Lu, Z.; Song, X.; Jiang, H.; et al. PTEN antagonises Tcl1/hnRNPK-mediated G6PD pre-mRNA splicing which contributes to hepatocarcinogenesis. Gut 2014, 63, 1635–1647. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, S.; Zhao, X.; Guo, H.; Ling, X.; Wang, L.; Fan, C.; Yu, J.; Zhou, S. OCT3 and SOX2 promote the transformation of Barrett’s esophagus to adenocarcinoma by regulating the formation of tumor stem cells. Oncol. Rep. 2014, 31, 1745–1753. [Google Scholar] [CrossRef] [Green Version]
- Herling, M.; Khoury, J.D.; Washington, L.B.T.; Duvic, M.; Keating, M.J.; Jones, D. A systematic approach to diagnosis of mature T-cell leukemias reveals heterogeneity among WHO categories. Blood 2004, 104, 328–335. [Google Scholar] [CrossRef] [Green Version]
- Narducci, M.G.; Pescarmona, E.; Lazzeri, C.; Signoretti, S.; Lavinia, A.M.; Remotti, D.; Scala, E.; Baroni, C.D.; Stoppacciaro, A.; Croce, C.M.; et al. Regulation of TCL1 expression in B- and T-cell lymphomas and reactive lymphoid tissues. Cancer Res. 2000, 60, 2095–2100. [Google Scholar]
- Nakayama, I.; Murao, S.; Kitazawa, S.; Azumi, A.; Yamamoto, M.; Maeda, S. Activation of the TCL1 protein in B cell lymphomas. Pathol. Int. 2000, 50, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Rodig, S.J.; Vergilio, J.A.; Shahsafaei, A.; Dorfman, D.M. Characteristic expression patterns of TCL1, CD38, and CD44 identify aggressive lymphomas harboring a MYC translocation. Am. J. Surg. Pathol. 2008, 32, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.X.; Li, S.J.; Niu, J.; Ma, Z.P.; Nuerlan, A.; Xue, J.; Wang, M.B.; Cui, W.L.; Abulajiang, G.; Sang, W.; et al. TCL1 as a hub protein associated with the PI3K/AKT signaling pathway in diffuse large B-cell lymphoma based on proteomics methods. Pathol. Res. Pract. 2020, 216, 152799. [Google Scholar] [CrossRef] [PubMed]
- Gualco, G.; Weiss, L.M.; Harrington, W.J.; Bacchi, C.E. Nodal diffuse large B-cell lymphomas in children and adolescents: Immunohistochemical expression patterns and c-MYC translocation in relation to clinical outcome. Am. J. Surg. Pathol. 2009, 33, 1815–1822. [Google Scholar] [CrossRef]
- Pescarmona, E.; Remotti, D.; Perez, M.; Monaco, S.; Pacchiarotti, A.; Faraggiana, T.; Russo, G.; Baroni, C.D. Expression of TCL1 and CD27 in primary cutaneous B-cell lymphomas. Histopathology 2006, 49, 343–348. [Google Scholar] [CrossRef]
- Lemal, R.; Bard-Sorel, S.; Montrieul, L.; Bay, J.O.; Ravinet, A.; Ledoux-Pilon, A.; Cagnard, N.; Bailly, S.; Morel, P.; Charlotte, F.; et al. TCL1 expression patterns in Waldenström macroglobulinemia. Mod. Pathol. 2016, 29, 83–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trinh, D.T.; Shibata, K.; Hirosawa, T.; Umezu, T.; Mizuno, M.; Kajiyama, H.; Kikkawa, F. Diagnostic utility of CD117, CD133, SALL4, OCT4, TCL1 and glypican-3 in malignant germ cell tumors of the ovary. J. Obstet. Gynaecol. Res. 2012, 38, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Staber, P.B.; Herling, M.; Bellido, M.; Jacobsen, E.D.; Davids, M.S.; Kadia, T.M.; Shustov, A.; Tournilhac, O.; Bachy, E.; Zaja, F.; et al. Consensus criteria for diagnosis, staging, and treatment response assessment of T-cell prolymphocytic leukemia. Blood 2019, 134, 1132–1143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diwan, A.H.; Prieto, V.G.; Herling, M.; Duvic, M.; Jone, D. Primary Sézary Syndrome Commonly Shows Low-Grade Cytologic Atypia and an Absence of Epidermotropism. Am. J. Clin. Pathol. 2005, 123, 510–515. [Google Scholar] [CrossRef]
- Herling, M.; Valbuena, J.R.; Jones, D.; Medeiros, L.J. Skin involvement in T-cell prolymphocytic leukemia. J. Am. Acad. Dermatol. 2007, 57, 533–534. [Google Scholar] [CrossRef] [PubMed]
- Valbuena, J.R.; Herling, M.; Admirand, J.H.; Padula, A.; Jones, D.; Medeiros, L.J. T-Cell Prolymphocytic Leukemia Involving Extramedullary Sites. Am. J. Clin. Pathol. 2005, 123, 456–464. [Google Scholar] [CrossRef]
- Bresin, A.; Ragone, G.; Cristofoletti, C.; Arcelli, D.; Bassi, C.; Caprini, E.; Fiorenza, M.T.; Citterich, M.H.; Russo, G.; Narducci, M.G. T Cell Leukemia/Lymphoma 1A is essential for mouse epidermal keratinocytes proliferation promoted by insulin-like growth factor 1. PLoS ONE 2018, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishimura, K.; Aizawa, S.; Nugroho, F.L.; Shiomitsu, E.; Tran, Y.T.H.; Bui, P.L.; Borisova, E.; Sakuragi, Y.; Takada, H.; Kurisaki, A.; et al. A Role for KLF4 in Promoting the Metabolic Shift via TCL1 during Induced Pluripotent Stem Cell Generation. Stem Cell Rep. 2017, 8, 787–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oberbeck, S.; Schrader, A.; Warner, K.; Jungherz, D.; Crispatzu, G.; von Jan, J.; Chmielewski, M.; Ianevski, A.; Diebner, H.H.; Mayer, P.; et al. Noncanonical effector functions of the T-memory–like T-PLL cell are shaped by cooperative TCL1A and TCR signaling. Blood 2020, 136, 2786–2802. [Google Scholar] [CrossRef] [PubMed]
- Auguin, D.; Barthe, P.; Royer, C.; Stern, M.H.; Noguchi, M.; Arold, S.T.; Roumestand, C. Structural basis for the co-activation of protein kinase B by T-cell leukemia-1 (TCL1) family proto-oncoproteins. J. Biol. Chem. 2004, 279, 35890–35902. [Google Scholar] [CrossRef] [Green Version]
- Palamarchuk, A.; Yan, P.S.; Zanesi, N.; Wang, L.; Rodrigues, B.; Murphy, M.; Balatti, V.; Bottoni, A.; Nazaryan, N.; Alder, H.; et al. Tcl1 protein functions as an inhibitor of de novo DNA methylation in B-cell chronic lymphocytic leukemia (CLL). Proc. Natl. Acad. Sci. USA 2012, 109, 2555–2560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Upchurch, G.M.; Haney, S.L.; Opavsky, R. Aberrant Promoter Hypomethylation in CLL: Does It Matter for Disease Development? Front. Oncol. 2016, 6, 182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Künstle, G.; Laine, J.; Pierron, G.; Kagami, S.; Nakajima, H.; Hoh, F.; Roumestand, C.; Stern, M.-H.; Noguchi, M. Identification of Akt Association and Oligomerization Domains of the Akt Kinase Coactivator TCL1. Mol. Cell. Biol. 2002, 22, 1513–1525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pekarsky, Y. Tcl1 enhances Akt kinase activity and mediates its nuclear translocation. Proc. Natl. Acad. Sci. USA 2000, 97, 3028–3033. [Google Scholar] [CrossRef]
- Noguchi, M.; Ropars, V.; Roumestand, C.; Suizu, F. Proto-oncogene TCL1: More than just a coactivator for Akt. FASEB J. 2007, 21, 2273–2284. [Google Scholar] [CrossRef] [PubMed]
- Laine, J.; Künstle, G.; Obata, T.; Sha, M.; Noguchi, M. The protooncogene TCL1 is an Akt kinase coactivator. Mol. Cell 2000, 6, 395–407. [Google Scholar] [CrossRef]
- Ragone, G.; Bresin, A.; Piermarini, F.; Lazzeri, C.; Picchio, M.C.; Remotti, D.; Kang, S.M.; Cooper, M.D.; Croce, C.M.; Narducci, M.G.; et al. The Tcl1 oncogene defines secondary hair germ cells differentiation at catagen-telogen transition and affects stem-cell marker CD34 expression. Oncogene 2009, 28, 1329–1338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matoba, R.; Niwa, H.; Masui, S.; Ohtsuka, S.; Carter, M.G.; Sharov, A.A.; Ko, M.S.H. Dissecting Oct3/4-regulated gene networks in embryonic stem cells by expression profiling. PLoS ONE 2006, 1, e26. [Google Scholar] [CrossRef]
- Ema, M.; Mori, D.; Niwa, H.; Hasegawa, Y.; Yamanaka, Y.; Hitoshi, S.; Mimura, J.; Kawabe, Y.-i.; Hosoya, T.; Morita, M.; et al. Krüppel-like factor 5 Is Essential for Blastocyst Development and the Normal Self-Renewal of Mouse ESCs. Cell Stem Cell 2008, 3, 555–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanova, N.; Dobrin, R.; Lu, R.; Kotenko, I.; Levorse, J.; DeCoste, C.; Schafer, X.; Lun, Y.; Lemischka, I.R. Dissecting self-renewal in stem cells with RNA interference. Nature 2006, 442, 533–538. [Google Scholar] [CrossRef]
- French, S.W.; Dawson, D.W.; Chen, H.-W.; Rainey, R.N.; Sievers, S.A.; Balatoni, C.E.; Wong, L.; Troke, J.J.; Nguyen, M.T.N.; Koehler, C.M.; et al. The TCL1 oncoprotein binds the RNase PH domains of the PNPase exoribonuclease without affecting its RNA degrading activity. Cancer Lett. 2007, 248, 198–210. [Google Scholar] [CrossRef]
- Prinz, C.; Vasyutina, E.; Lohmann, G.; Schrader, A.; Romanski, S.; Hirschhäuser, C.; Mayer, P.; Frias, C.; Herling, C.D.; Hallek, M.; et al. Organometallic nucleosides induce non-classical leukemic cell death that is mitochondrial-ROS dependent and facilitated by TCL1-oncogene burden. Mol. Cancer 2015, 14, 114. [Google Scholar] [CrossRef] [Green Version]
- Noh, K.H.; Kim, B.W.; Song, K.H.; Cho, H.; Lee, Y.H.; Kim, J.H.; Chung, J.Y.; Kim, J.H.; Hewitt, S.M.; Seong, S.Y.; et al. Nanog signaling in cancer promotes stem-like phenotype and immune evasion. J. Clin. Investig. 2012, 122, 4077–4093. [Google Scholar] [CrossRef] [Green Version]
- Song, K.H.; Oh, S.J.; Kim, S.; Cho, H.; Lee, H.J.; Song, J.S.; Chung, J.Y.; Cho, E.; Lee, J.; Jeon, S.; et al. HSP90A inhibition promotes anti-tumor immunity by reversing multi-modal resistance and stem-like property of immune-refractory tumors. Nat. Commun. 2020, 11, 562. [Google Scholar] [CrossRef] [PubMed]
- Pekarsky, Y.; Zanesi, N.; Croce, C.M. Molecular basis of CLL. Semin. Cancer Biol. 2010, 20, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Rathmell, J.C.; Elstrom, R.L.; Cinalli, R.M.; Thompson, C.B. Activated Akt promotes increased resting T cell size, CD28-independent T cell growth, and development of autoimmunity and lymphoma. Eur. J. Immunol. 2003, 33, 2223–2232. [Google Scholar] [CrossRef]
- Gaudio, E.; Spizzo, R.; Paduano, F.; Luo, Z.; Efanov, A.; Palamarchuk, A.; Leber, A.S.; Kaou, M.; Zanesi, N.; Bottoni, A.; et al. Tcl1 interacts with Atm and enhances NF-κB activation in hematologic malignancies. Blood 2012, 119, 180–187. [Google Scholar] [CrossRef] [Green Version]
- Motiwala, T.; Zanesi, N.; Datta, J.; Roy, S.; Kutay, H.; Checovich, A.M.; Kaou, M.; Zhong, Y.; Johnson, A.J.; Lucas, D.M.; et al. AP-1 elements and TCL1 protein regulate expression of the gene encoding protein tyrosine phosphatase PTPROt in leukemia. Blood 2011, 118, 6132–6140. [Google Scholar] [CrossRef] [Green Version]
- Woo, S.R.; Lee, H.J.; Oh, S.J.; Kim, S.; Park, S.H.; Lee, J.; Song, K.H.; Kim, T.W. Stabilization of HDAC1 via TCL1-pAKT-CHFR axis is a key element for NANOG-mediated multi-resistance and stem-like phenotype in immune-edited tumor cells. Biochem. Biophys. Res. Commun. 2018, 503, 1812–1818. [Google Scholar] [CrossRef]
- Jiang, J.; Chan, Y.S.; Loh, Y.H.; Cai, J.; Tong, G.Q.; Lim, C.A.; Robson, P.; Zhong, S.; Ng, H.H. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 2008, 10, 353–360. [Google Scholar] [CrossRef]
- Doerr, J.R.; Malone, C.S.; Fike, F.M.; Gordon, M.S.; Soghomonian, S.V.; Thomas, R.K.; Tao, Q.; Murray, P.G.; Diehl, V.; Teitell, M.A.; et al. Patterned CpG methylation of silenced B cell gene promoters in classical hodgkin lymphoma-derived and primary effusion lymphoma cell lines. J. Mol. Biol. 2005, 350, 631–640. [Google Scholar] [CrossRef]
- Kuraishy, A.I.; French, S.W.; Sherman, M.; Herling, M.; Jones, D.; Wall, R.; Teitell, M.A. TORC2 regulates germinal center repression of the TCL1 oncoprotein to promote B cell development and inhibit transformation. Proc. Natl. Acad. Sci. USA 2007, 104, 10175–10180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiromura, M.; Suizu, F.; Narita, M.; Kinowaki, K.; Noguchi, M. Identification of nerve growth factor-responsive element of the TCL1 promoter as a novel negative regulatory element. J. Biol. Chem. 2006, 281, 27753–27764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anastasiadou, E.; Boccellato, F.; Vincenti, S.; Rosato, P.; Bozzoni, I.; Frati, L.; Faggioni, A.; Presutti, C.; Trivedi, P. Epstein-Barr virus encoded LMP1 downregulates TCL1 oncogene through miR-29b. Oncogene 2010, 29, 1316–1328. [Google Scholar] [CrossRef] [Green Version]
- Boccellato, F.; Anastasiadou, E.; Rosato, P.; Kempkes, B.; Frati, L.; Faggioni, A.; Trivedi, P. EBNA2 Interferes with the Germinal Center Phenotype by Downregulating BCL6 and TCL1 in Non-Hodgkin’s Lymphoma Cells. J. Virol. 2007, 81, 2274–2282. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Sakakibara, S.; Maruo, S.; Zhao, B.; Calderwood, M.A.; Holthaus, A.M.; Lai, C.-Y.; Takada, K.; Kieff, E.; Johannsen, E. Epstein-Barr Virus Nuclear Protein 3C Domains Necessary for Lymphoblastoid Cell Growth: Interaction with RBP-Jκ Regulates TCL1. J. Virol. 2009, 83, 12368–12377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balatti, V.; Rizzotto, L.; Miller, C.; Palamarchuk, A.; Fadda, P.; Pandolfo, R.; Rassenti, L.Z.; Hertlein, E.; Ruppert, A.S.; Lozanski, A.; et al. TCL1 targeting miR-3676 is codeleted with tumor protein p53 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2015, 112, 2169–2174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardinaud, B.; Moreilhon, C.; Marcet, B.; Robbe-Sermesant, K.; LeBrigand, K.; Mari, B.; Eclache, V.; Cymbalista, F.; Raynaud, S.; Barbry, P. miR-34b/miR-34c: A regulator of TCL1 expression in 11q- chronic lymphocytic leukaemia? Leukemia 2009, 23, 2174–2177. [Google Scholar] [CrossRef]
- Gaudio, E.; Paduano, F.; Ngankeu, A.; Lovat, F.; Fabbri, M.; Sun, H.L.; Gasparini, P.; Efanov, A.; Peng, Y.; Zanesi, N.; et al. Heat shock protein 70 regulates Tcl1 expression in leukemia and lymphomas. Blood 2013, 121, 351–359. [Google Scholar] [CrossRef] [Green Version]
- French, S.W.; Malone, C.S.; Shen, R.R.; Renard, M.; Henson, S.E.; Miner, M.D.; Wall, R.; Teitell, M.A. Sp1 transactivation of the TCL1 oncogene. J. Biol. Chem. 2003, 278, 948–955. [Google Scholar] [CrossRef] [Green Version]
- Voltan, R.; Di Iasio, M.G.; Bosco, R.; Valeri, N.; Pekarski, Y.; Tiribelli, M.; Secchiero, P.; Zauli, G. Nutlin-3 downregulates the expression of the oncogene TCL1 in primary B chronic lymphocytic leukemic cells. Clin. Cancer Res. 2011, 17, 5649–5655. [Google Scholar] [CrossRef] [Green Version]
- Shen, R.R.; Ferguson, D.O.; Renard, M.; Hoyer, K.K.; Kim, U.; Hao, X.; Alt, F.W.; Roeder, R.G.; Morse, H.C.; Teitell, M.A. Dysregulated TCL1 requires the germinal center and genome instability for mature B-cell transformation. Blood 2006, 108, 1991–1998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, H.S.; Cisse, B.; Bunin, A.; Lewis, K.L.; Reizis, B. Continuous Expression of the Transcription Factor E2-2 Maintains the Cell Fate of Mature Plasmacytoid Dendritic Cells. Immunity 2010, 33, 905–916. [Google Scholar] [CrossRef] [Green Version]
- Ceribelli, M.; Hou, Z.E.; Kelly, P.N.; Huang, D.W.; Wright, G.; Ganapathi, K.; Evbuomwan, M.O.; Pittaluga, S.; Shaffer, A.L.; Marcucci, G.; et al. A Druggable TCF4- and BRD4-Dependent Transcriptional Network Sustains Malignancy in Blastic Plasmacytoid Dendritic Cell Neoplasm. Cancer Cell 2016, 30, 764–778. [Google Scholar] [CrossRef] [Green Version]
- Renosi, F.; Roggy, A.; Giguelay, A.; Soret, L.; Viailly, P.J.; Cheok, M.; Biichle, S.; Angelot-Delettre, F.; Asnafi, V.; Macintyre, E.; et al. Transcriptomic and genomic heterogeneity in blastic plasmacytoid dendritic cell neoplasms: From ontogeny to oncogenesis. Blood Adv. 2021, 5, 1540–1551. [Google Scholar] [CrossRef] [PubMed]
- Ingle, J.N.; Schaid, D.J.; Goss, P.E.; Liu, M.; Mushiroda, T.; Chapman, J.A.W.; Kubo, M.; Jenkins, G.D.; Batzler, A.; Shepherd, L.; et al. Genome-wide associations and functional genomic studies of musculoskeletal adverse events in women receiving aromatase inhibitors. J. Clin. Oncol. 2010, 28, 4674–4682. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Wang, L.; Bongartz, T.; Hawse, J.R.; Markovic, S.N.; Schaid, D.J.; Mushiroda, T.; Kubo, M.; Nakamura, Y.; Kamatani, N.; et al. Aromatase inhibitors, estrogens and musculoskeletal pain: Estrogen-dependent T-cell leukemia 1A (TCL1A) gene-mediated regulation of cytokine expression. Breast Cancer Res. 2012, 14, R41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hertz, D.L.; Smith, K.L.; Zong, Y.; Gersch, C.L.; Pesch, A.M.; Lehman, J.; Blackford, A.L.; Henry, N.L.; Kidwell, K.M.; Rae, J.M.; et al. Further Evidence That OPG rs2073618 Is Associated With Increased Risk of Musculoskeletal Symptoms in Patients Receiving Aromatase Inhibitors for Early Breast Cancer. Front. Genet. 2021, 12, 662734. [Google Scholar] [CrossRef]
- Efanov, A.; Zanesi, N.; Nazaryan, N.; Santanam, U.; Palamarchuk, A.; Croce, C.M.; Pekarsky, Y. CD5+CD23+ leukemic cell populations in TCL1 transgenic mice show significantly increased proliferation and Akt phosphorylation. Leukemia 2010, 24, 970–975. [Google Scholar] [CrossRef] [PubMed]
- Pekarsky, Y.; Santanam, U.; Cimmino, A.; Palamarchuk, A.; Efanov, A.; Maximov, V.; Volinia, S.; Alder, H.; Liu, C.G.; Rassenti, L.; et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006, 66, 11590–11593. [Google Scholar] [CrossRef] [Green Version]
- Seifert, M.; Sellmann, L.; Bloehdorn, J.; Wein, F.; Stilgenbauer, S.; Dürig, J.; Küppers, R. Cellular origin and pathophysiology of chronic lymphocytic leukemia. J. Exp. Med. 2012, 209, 2183–2198. [Google Scholar] [CrossRef] [Green Version]
- Frezzato, F.; Raggi, F.; Martini, V.; Severin, F.; Trimarco, V.; Visentin, A.; Scomazzon, E.; Accordi, B.; Bresolin, S.; Piazza, F.; et al. HSP70/HSF1 axis, regulated via a PI3K/AKT pathway, is a druggable target in chronic lymphocytic leukemia. Int. J. Cancer 2019, 145, 3089–3100. [Google Scholar] [CrossRef]
- Sivina, M.; Hartmann, E.; Vasyutina, E.; Boucas, J.M.; Breuer, A.; Keating, M.J.; Wierda, W.G.; Rosenwald, A.; Herling, M.; Burger, J.A. Stromal cells modulate TCL1 expression, interacting AP-1 components and TCL1-targeting micro-RNAs in chronic lymphocytic leukemia. Leukemia 2012, 26, 1812–1820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiss, C.; Nishikawa, J.; Takada, K.; Trivedi, P.; Klein, G.; Szekely, L. T cell leukemia I oncogene expression depends on the presence of Epstein-Barr virus in the virus-carrying Burkitt lymphoma lines. Proc. Natl. Acad. Sci. USA 2003, 100, 4813–4818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bell, A.; Rickinson, A.B. Epstein–Barr virus, the TCL-1 oncogene and Burkitt’s lymphoma. Trends Microbiol. 2003, 11, 495–497. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Vaeth, S.; Cuomo, L.; Boccellato, F.; Vincenti, S.; Cirone, M.; Presutti, C.; Junker, S.; Winberg, G.; Frati, L.; et al. Epstein-Barr virus infection leads to partial phenotypic reversion of terminally differentiated malignant B cells. Cancer Lett. 2009, 284, 165–174. [Google Scholar] [CrossRef]
- Bajaj, B.G.; Murakami, M.; Robertson, E.S. Molecular biology of EBV in relationship to AIDS-associated oncogenesis. Cancer Treat. Res. 2007, 133, 141–162. [Google Scholar] [CrossRef]
- Cohen, J.I.; Bollard, C.M.; Khanna, R.; Pittaluga, S. Current understanding of the role of Epstein-Barr virus in lymphomagenesis and therapeutic approaches to EBV-associated lymphomas. Leuk. Lymphoma 2008, 49, 27–34. [Google Scholar] [CrossRef] [Green Version]
- Mancao, C.; Hammerschmidt, W. Epstein-Barr virus latent membrane protein 2A is a B-cell receptor mimic and essential for B-cell survival. Blood 2007, 110, 3715–3721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trivedi, P.; Takazawa, K.; Zompetta, C.; Cuomo, L.; Anastasiadou, E.; Carbone, A.; Uccini, S.; Belardelli, F.; Takada, K.; Frati, L.; et al. Infection of HHV-8+ primary effusion lymphoma cells with a recombinant Epstein-Barr virus leads to restricted EBV latency, altered phenotype, and increased tumorigenicity without affecting TCL1 expression. Blood 2004, 103, 313–316. [Google Scholar] [CrossRef] [Green Version]
- Teitell, M.A.; Lones, M.A.; Perkins, S.L.; Sanger, W.G.; Cairo, M.S.; Said, J.W. TCL1 Expression and Epstein-Barr Virus Status in Pediatric Burkitt Lymphoma. Am. J. Clin. Pathol. 2005, 124, 569–575. [Google Scholar] [CrossRef]
- Pagano, L.; Valentini, C.G.; Grammatico, S.; Pulsoni, A. Blastic plasmacytoid dendritic cell neoplasm: Diagnostic criteria and therapeutical approaches. Br. J. Haematol. 2016, 174, 188–202. [Google Scholar] [CrossRef]
- Brinas, F.; Danger, R.; Brouard, S. TCL1A, B Cell Regulation and Tolerance in Renal Transplantation. Cells 2021, 10, 1367. [Google Scholar] [CrossRef] [PubMed]
- Petock, J.M.; Torshin, I.Y.; Wang, Y.-F.; Du Bois, G.C.; Croce, C.M.; Harrison, R.W.; Weber, I.T. Crystal Structures of Tcl1 Family Oncoproteins and Their Conserved Surface Features. Sci. World J. 2002, 2, 1876–1884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, J.; Rawal, S.; Chu, F.; Park, H.J.; Sharma, R.; Delgado, D.A.; Fayad, L.; Fanale, M.; Romaguera, J.; Luong, A.; et al. TCL1: A shared tumor-associated antigen for immunotherapy against B-cell lymphomas. Blood 2012, 120, 1613–1623. [Google Scholar] [CrossRef] [Green Version]
- Weng, J.; Moriarty, K.; Pan, Y.; MA, M.C.J.; Mathur, R.; Zhang, Z.; Torikai, H.; Maiti, S.N.; Chu, F.; Cheng, X.; et al. Adoptive T-Cell Therapy with TCL1-Specific TCR for B-Cell Lymphomas. Blood 2018, 132, 3488. [Google Scholar] [CrossRef]
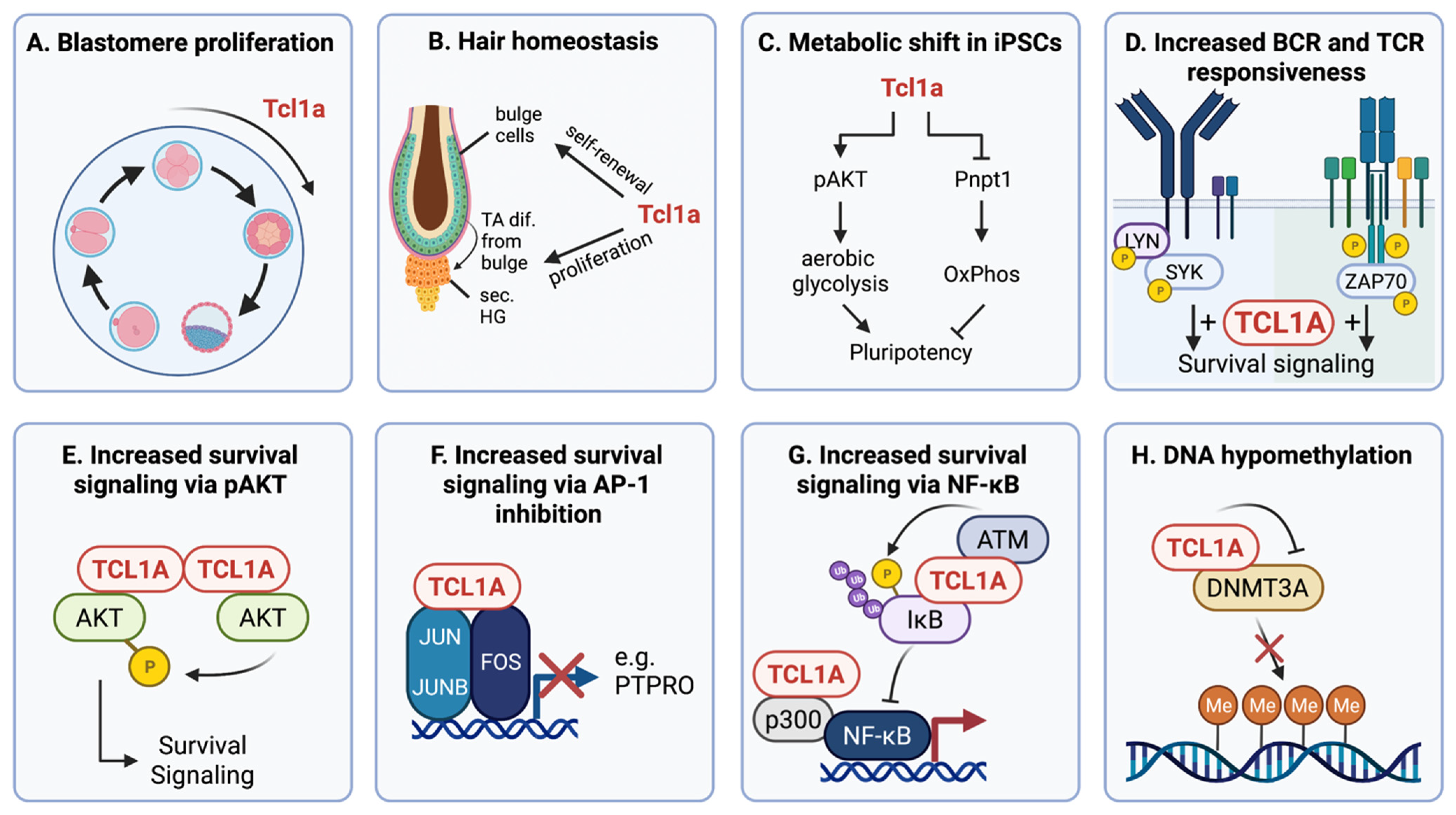
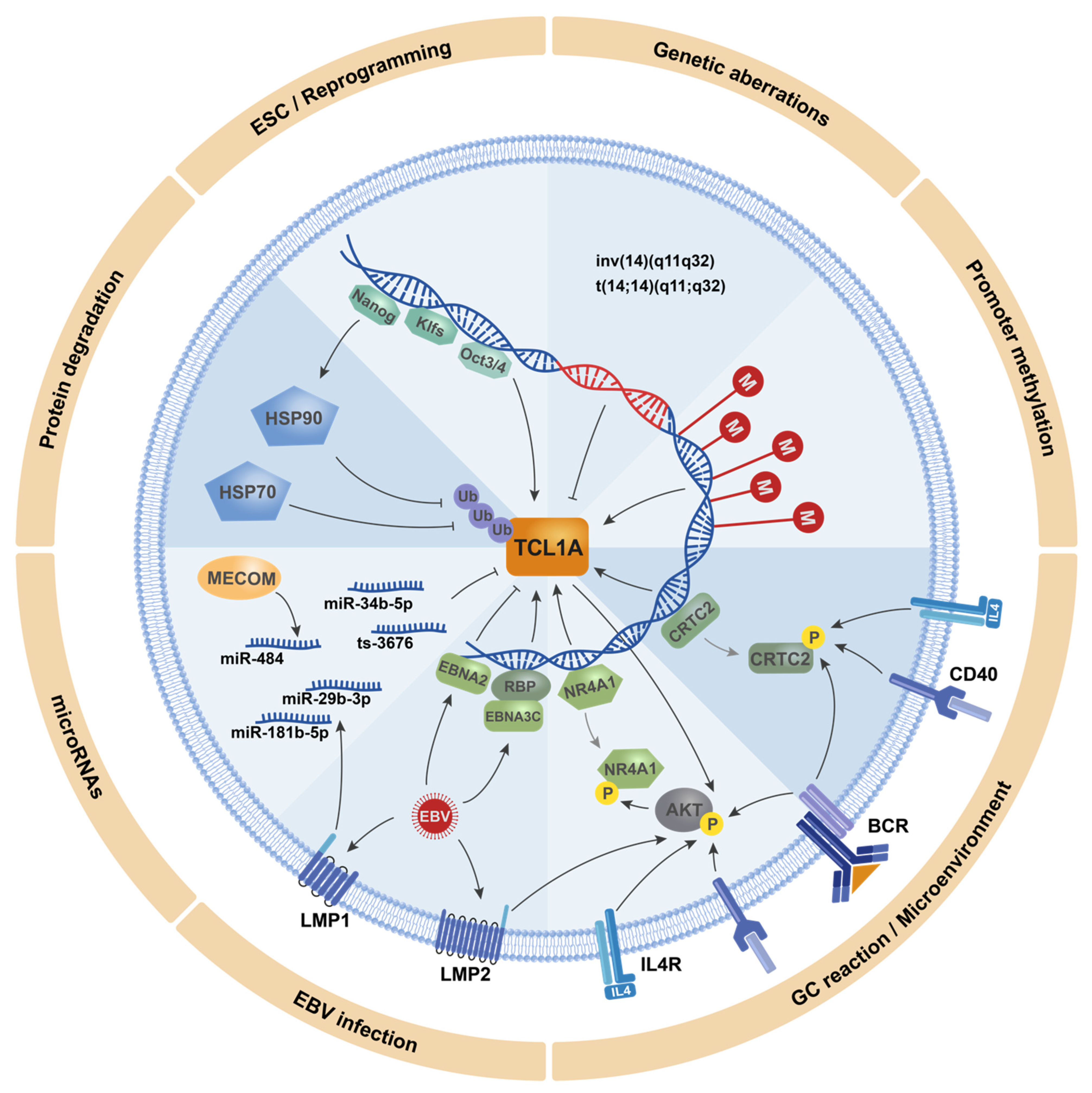
| Entity | N | TCL1A Expression [% Cases] | Reference | Prognostic Implications |
|---|---|---|---|---|
| Leukemia/Lymphoma | ||||
| T-cell leukemias/lymphomas | ||||
| T-cell prolymphocytic leukemia | 38–59 | 71–75 | [32,33,46] | Shorter OS [26] |
| T-(acute) lymphoblastic leukemia/lymphoma | 7–47 | 14–36 | [32,33] | - |
| Adult T-cell leukemia/lymphoma | 5 | 20 | [32] | - |
| B-cell leukemias/lymphomas | ||||
| B-(acute) lymphoblastic leukemia/lymphoma | 4–55 | 75–85 | [12,47] | - |
| Mantle cell lymphoma | 5–58 | 84–100 | [12,47,48] | Shorter LSS [38] |
| Burkitt Lymphoma | 5–16 | 94–100 | [12,47,49] | - |
| Follicular lymphoma | 11–49 | 57–75 | [12,47,48] | - |
| Diffuse large B-cell lymphoma | 11–15 | 18–60 | [12,47,48,49] | Shorter OS [50] |
| Pediatric diffuse large B-cell lymphoma | 16 | 31 | [51] | - |
| CD5- Lymphoproliferative disorder | 2 | 100 | [12] | - |
| MALT lymphoma | 9–23 | 11–83 | [12,48] | - |
| Lymphoplasmacytic lymphoma | 4 | 75 | [48] | - |
| Small lymphocytic lymphoma | 2 | 100 | [48] | - |
| Chronic lymphocytic leukemia | 11–126 | 90–100 | [28,47] | Shorter TFS/PFS [24,38] |
| Cutaneous B-cell lymphoma | 9–25 | 20–55 | [47,52] | - |
| Waldenström macroglobulinemia | 57 | 79 | [53] | No [53] |
| Myeloid neoplasms | ||||
| Extramedullary myeloid cell tumors | 14 | 7 | [32] | - |
| Blastic plasmacytoid dendritic cell neoplasm | 12–91 | 83–99 | [32,35] | - |
| Solid tumors | ||||
| Bladder cancer | 10 | 40 | [42] | - |
| Prostate cancer | 5 | 80 | [42] | - |
| Colon cancer | 5 | 60 | [42] | - |
| Colorectal cancer | 278 | >70 | [30] | Shorter DSS [30] |
| Hepatocellular carcinoma | 65 | >44 | [44] | Shorter OS [44] |
| Germ cell tumors | ||||
| Classical seminoma | 13–55 | 77–100 | [9,39,40] | - |
| Embryonal carcinoma | 34 | 9 | [40] | - |
| Intratubular germ cell neoplasia | 40–50 | 100 | [39,40] | - |
| Spermatocytic seminoma | 6 | 17 | [40] | - |
| Ovarian dysgerminoma | 25 | 100 | [54] | - |
| Ovarian yolk sac tumor | 29 | 59 | [54] | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stachelscheid, J.; Jiang, Q.; Herling, M. The Modes of Dysregulation of the Proto-Oncogene T-Cell Leukemia/Lymphoma 1A. Cancers 2021, 13, 5455. https://doi.org/10.3390/cancers13215455
Stachelscheid J, Jiang Q, Herling M. The Modes of Dysregulation of the Proto-Oncogene T-Cell Leukemia/Lymphoma 1A. Cancers. 2021; 13(21):5455. https://doi.org/10.3390/cancers13215455
Chicago/Turabian StyleStachelscheid, Johanna, Qu Jiang, and Marco Herling. 2021. "The Modes of Dysregulation of the Proto-Oncogene T-Cell Leukemia/Lymphoma 1A" Cancers 13, no. 21: 5455. https://doi.org/10.3390/cancers13215455






