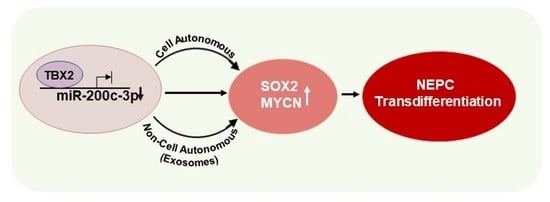
TBX2 Drives Neuroendocrine Prostate Cancer through Exosome-Mediated Repression of miR-200c-3p
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatments
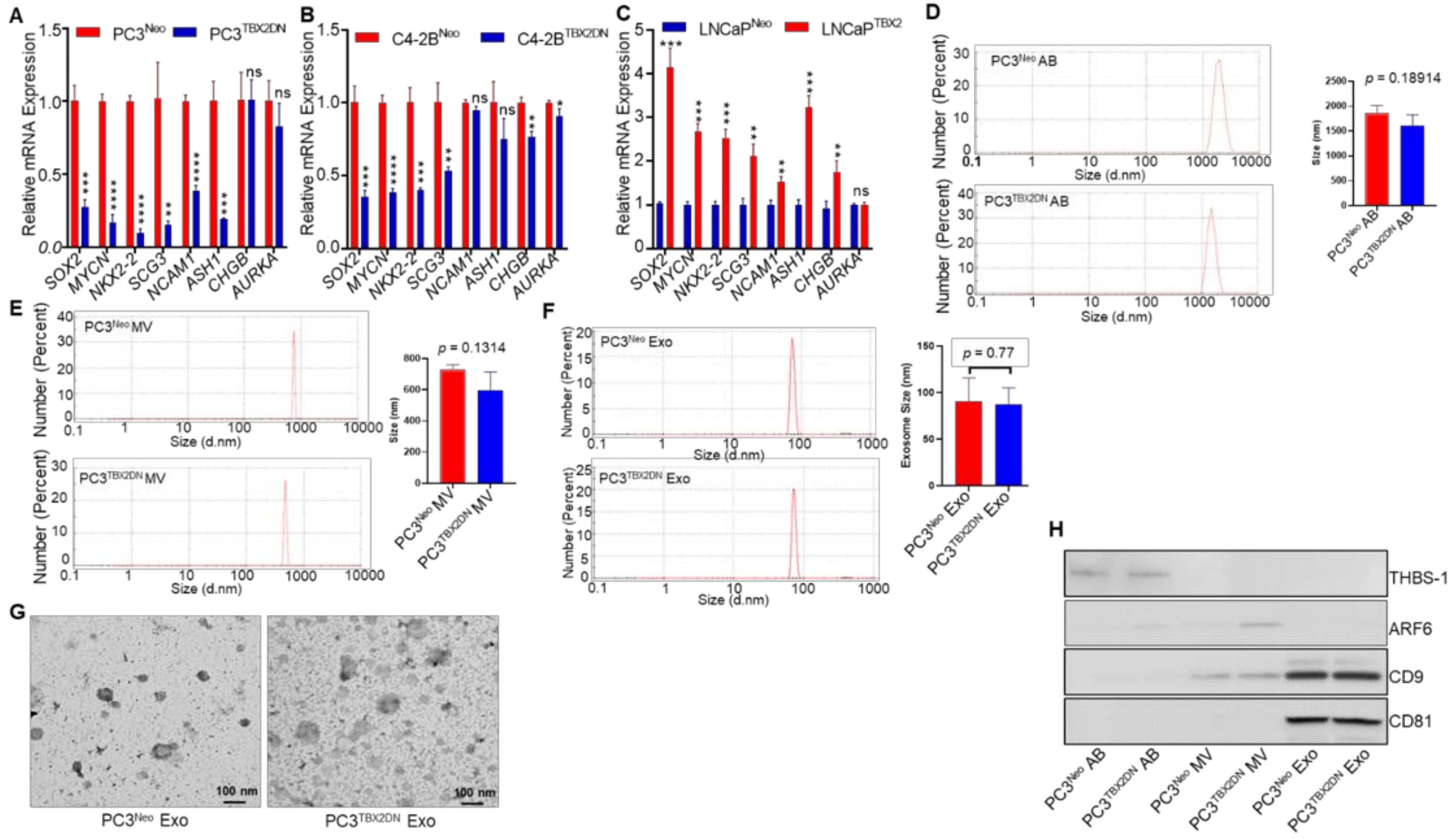
2.2. Exosome Isolation and Characterization
2.3. RNA Isolation, cDNA Synthesis, and quantitative real-time RT-PCR (qRT-PCR) Analysis
2.4. Protein Extraction and Quantification
2.5. SDS-PAGE and Western Blot Analysis
2.6. In Silico Analysis for microRNA Putative Targets
2.7. Plasmid Propagation, Transduction, and Modulation of miR-200c-3p Expression
2.8. Chromatin Immunoprecipitation (ChIP) Assays
2.9. Tumor Xenograft Experiments in Mice
2.10. Immunohistochemistry of Tumor Tissues
2.11. Exosome Internalization and Fluorescence Microscopy
2.12. Statistical Analysis
3. Results
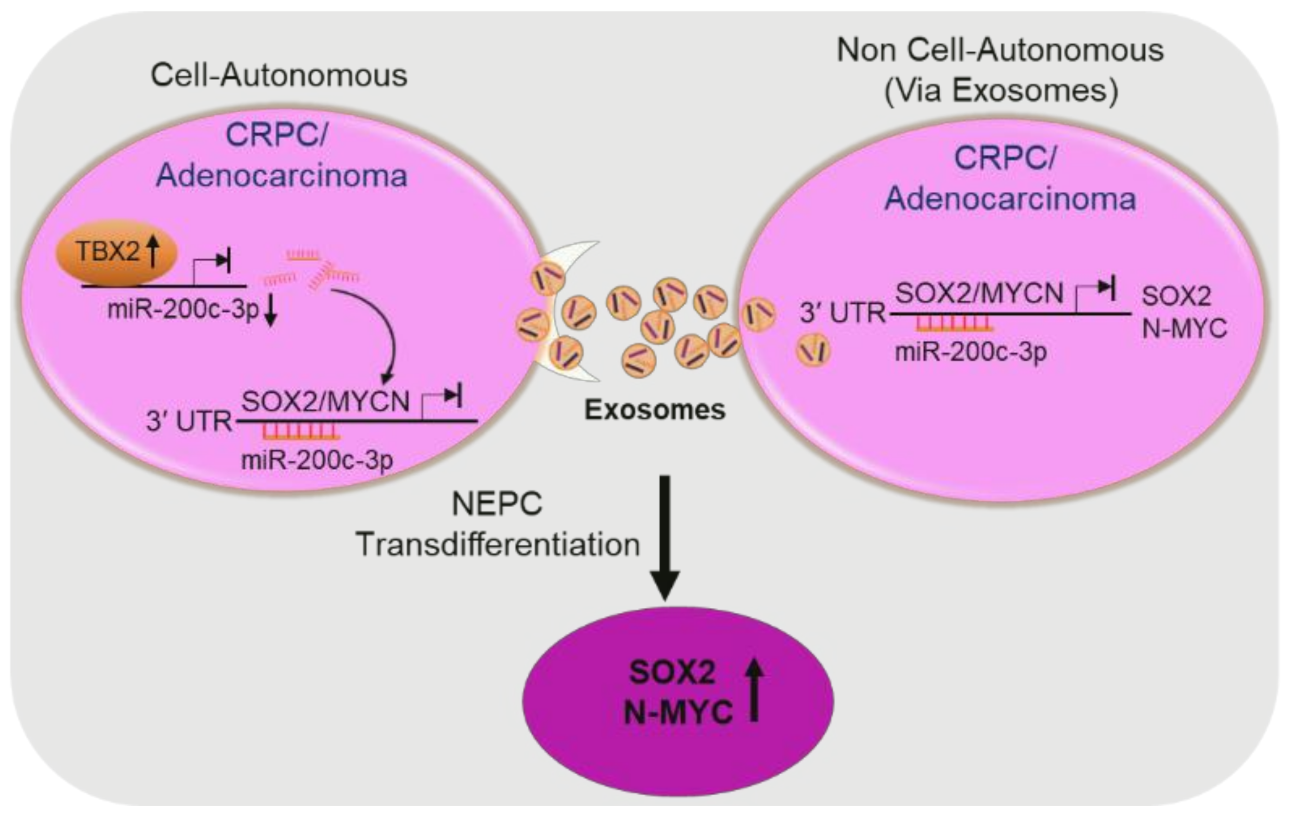
3.1. TBX2 Regulates Expression of NEPC Markers in PCa via Cell-Autonomous and Exosome-Mediated Non Cell-Autonomous Mechanisms
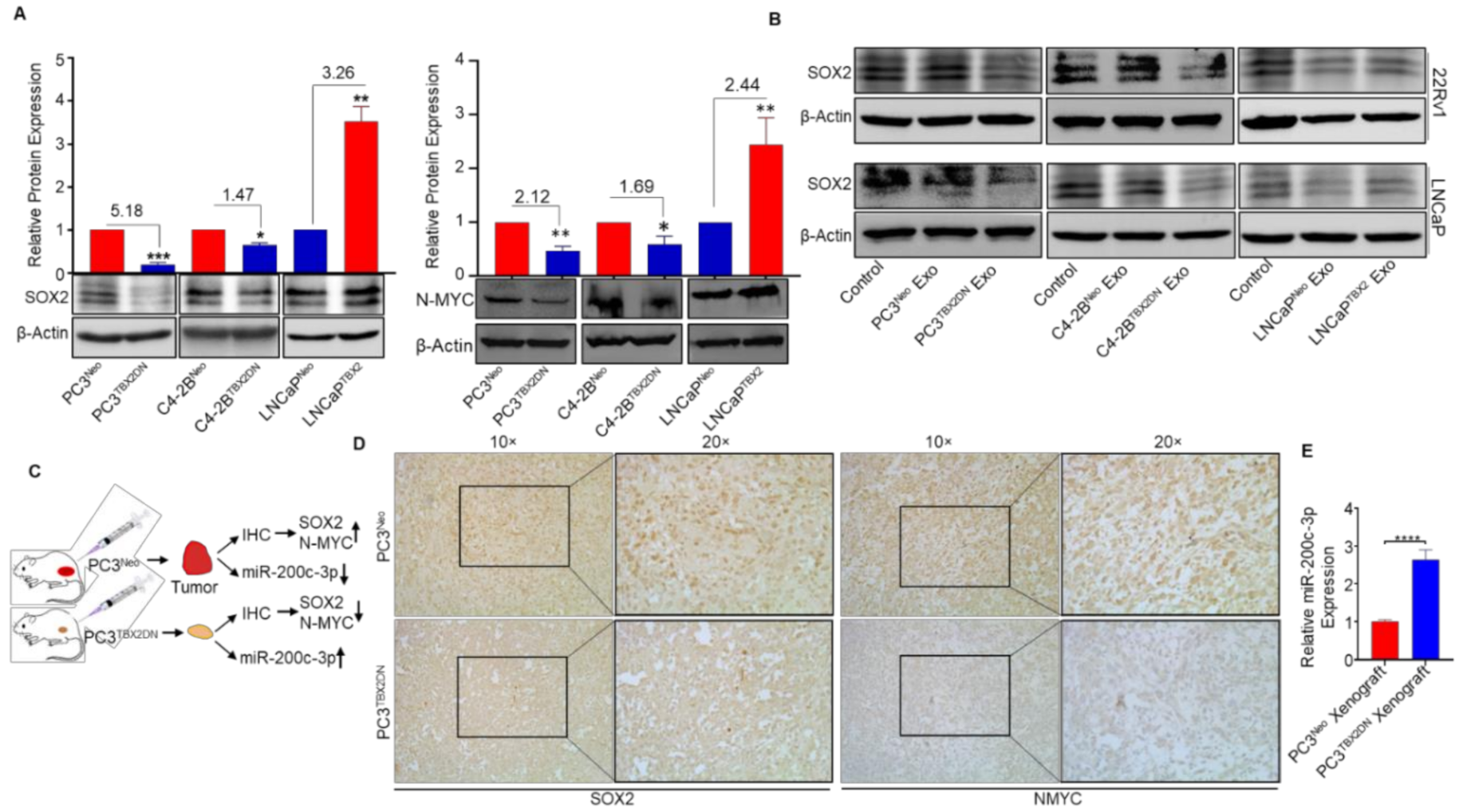
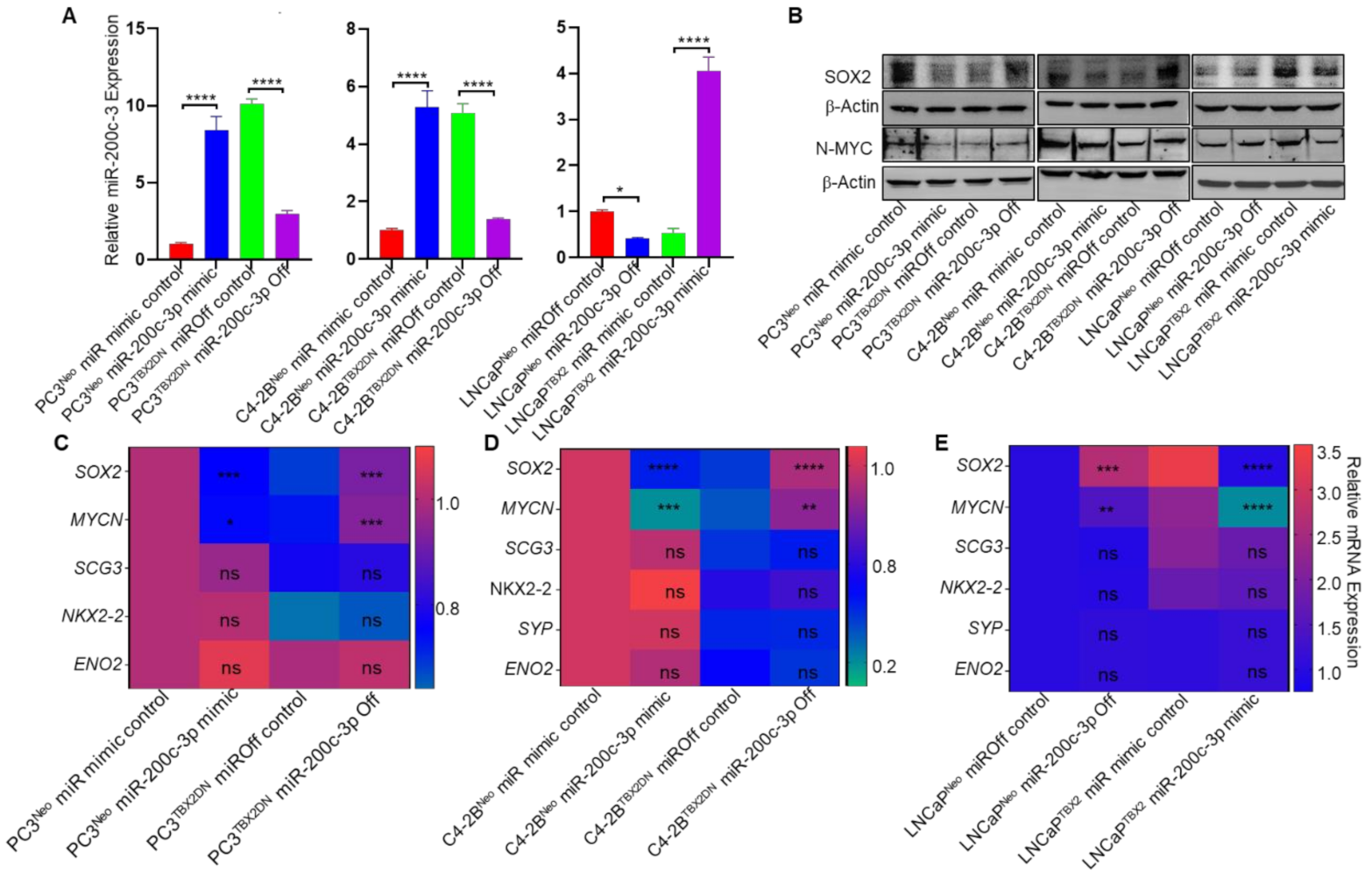
3.2. miR-200c-3p Is Downstream of TBX2 Signaling in PCa
3.3. Abrogated Metastasis in a Mouse Model Caused by Blocking Endogenous TBX2 Results in the Upregulation of miR-200c-3p and Downregulation of SOX2/N-MYC
3.4. miR-200c-3p Is the Intermediary Effector in TBX2 Regulation of SOX2 and MYCN
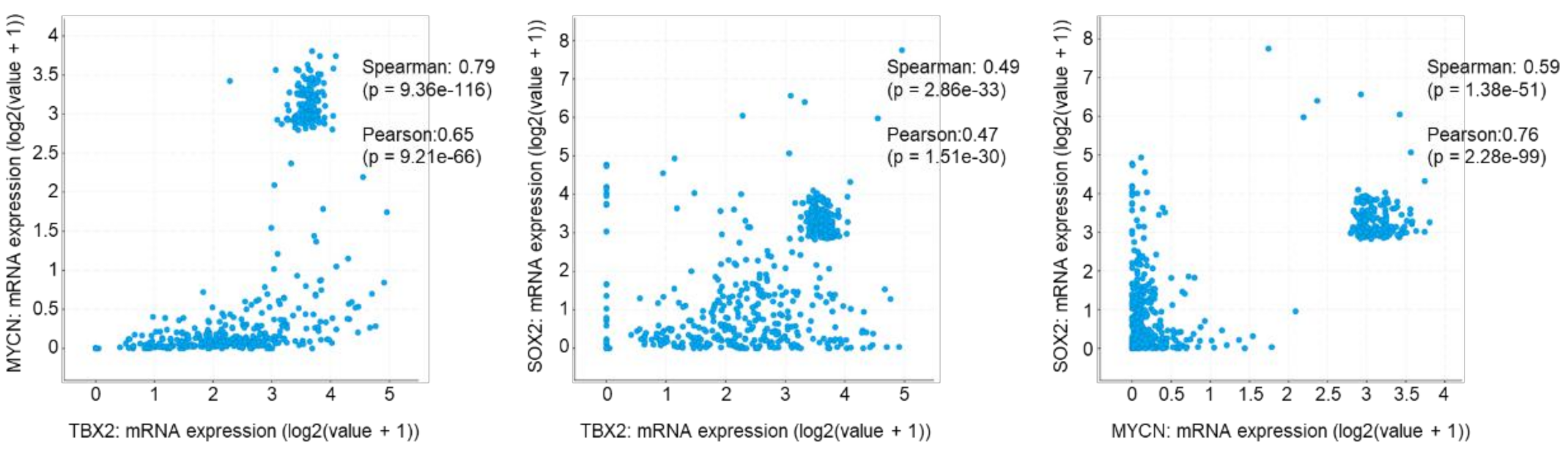
3.5. TBX2 Is Associated with SOX2 and MYCN in Human PCa
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AB/ABs | Apoptotic bodies |
| ANOVA | Analysis of Variance |
| ARF6 | ADP Ribosylation Factor 6 |
| ASH1 | Absent Small and Homeotic Disks Protein 1 Homolog |
| AURKA | Aurora Kinase A |
| C4-2BTBX2DN | Genetic modulation of endogenous TBX2 in C4-2B human prostate cancer cells using the Dominant Negative (DN) construct |
| CD | Cluster of Differentiation |
| CHG | Chromogranin |
| CRPC | Castrate Resistant Prostate Cancer |
| DAPI | 4,6-diamino-2-phenylindole |
| ENO2 | Enolase 2 |
| Exo | Exosomes |
| FITC | Fluorescein isothiocyanate |
| hsa-miR | human microRNA |
| IACUC | Institutional Animal Care and Use Committee |
| IgG | Immunoglobulin G |
| IHC | Immunohistochemistry |
| ires GFP | Internal Ribosome Entry Sites Green Fluorescence Protein |
| KEGG | Kyoto Encyclopaedia of Genes and Genomes |
| LNCaPTBX2 | Genetic modulation of TBX2 in LNCaP human prostate cancer cells using the over-expression construct |
| miR | microRNA |
| Ms | Mouse |
| MV/MVs | Microvesicles |
| NCAM1 | Neural cell adhesion molecule1 |
| NE | Neuroendocrine |
| Neo | Control Vector |
| NEPC | Neuroendocrine Prostate Cancer |
| NGS | Next Generation Sequencing |
| NKX2-2 | NK2 HomeoBox 2 |
| N-MYC (encoded by MYCN) | Neuroblastoma-Derived V-Myc Avian Myelocytomatosis Viral Related Oncogenic protein |
| PCa | Prostate Cancer |
| PC3TBX2DN | Genetic modulation of endogenous TBX2 in PC3 human prostate cancer cells using the Dominant Negative (DN) construct |
| qRT-PCR | Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction |
| RT | Room temperature |
| SCG3 | Secretogranin III |
| SD | Standard deviation |
| SF/SFs | Soluble factors |
| SOX2 | SRY-Box transcription Factor 2 |
| SYP | Synaptophysin |
| TBX2 | T-Box transcription Factor 2 |
| T.D | Transmitted Detector |
| THBS1 | Thrombospondin 1 |
| t-NEPC | Treatment-induced Neuroendocrine Prostate Cancer |
| UTR | UnTranslated Region |
References
- Patel, G.K.; Chugh, N.; Tripathi, M. Neuroendocrine Differentiation of Prostate Cancer-An Intriguing Example of Tumor Evolution at Play. Cancers 2019, 11, 1405. [Google Scholar] [CrossRef] [Green Version]
- Ku, S.Y.; Gleave, M.E.; Beltran, H. Towards precision oncology in advanced prostate cancer. Nat Rev. Urol. 2019, 16, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Beltran, H.; Tomlins, S.; Aparicio, A.; Arora, V.; Rickman, D.; Ayala, G.; Huang, J.; True, L.; Gleave, M.E.; Soule, H.; et al. Aggressive variants of castration-resistant prostate cancer. Clin. Cancer Res. 2014, 20, 2846–2850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlachostergios, P.J.; Puca, L.; Beltran, H. Emerging Variants of Castration-Resistant Prostate Cancer. Curr. Oncol. Rep. 2017, 19, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akamatsu, S.; Inoue, T.; Ogawa, O.; Gleave, M.E. Clinical and molecular features of treatment-related neuroendocrine prostate cancer. Int. J. Urol. 2018, 25, 345–351. [Google Scholar] [CrossRef] [Green Version]
- Yasumizu, Y.; Rajabi, H.; Jin, C.; Hata, T.; Pitroda, S.; Long, M.D.; Hagiwara, M.; Li, W.; Hu, Q.; Liu, S.; et al. MUC1-C regulates lineage plasticity driving progression to neuroendocrine prostate cancer. Nat. Commun. 2020, 11, 338. [Google Scholar] [CrossRef] [Green Version]
- Rotinen, M.; You, S.; Yang, J.; Coetzee, S.G.; Reis-Sobreiro, M.; Huang, W.C.; Huang, F.; Pan, X.; Yanez, A.; Hazelett, D.J.; et al. ONECUT2 is a targetable master regulator of lethal prostate cancer that suppresses the androgen axis. Nat. Med. 2018, 24, 1887–1898. [Google Scholar] [CrossRef]
- Bishop, J.L.; Thaper, D.; Vahid, S.; Davies, A.; Ketola, K.; Kuruma, H.; Jama, R.; Nip, K.M.; Angeles, A.; Johnson, F.; et al. The Master Neural Transcription Factor BRN2 Is an Androgen Receptor-Suppressed Driver of Neuroendocrine Differentiation in Prostate Cancer. Cancer Discov 2017, 7, 54–71. [Google Scholar] [CrossRef] [Green Version]
- Mu, P.; Zhang, Z.; Benelli, M.; Karthaus, W.R.; Hoover, E.; Chen, C.C.; Wongvipat, J.; Ku, S.Y.; Gao, D.; Cao, Z.; et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 2017, 355, 84–88. [Google Scholar] [CrossRef] [Green Version]
- Metz, E.P.; Wilder, P.J.; Dong, J.; Datta, K.; Rizzino, A. Elevating SOX2 in prostate tumor cells upregulates expression of neuroendocrine genes, but does not reduce the inhibitory effects of enzalutamide. J. Cell Physiol. 2020, 235, 3731–3740. [Google Scholar] [CrossRef]
- Lee, J.K.; Phillips, J.W.; Smith, B.A.; Park, J.W.; Stoyanova, T.; McCaffrey, E.F.; Baertsch, R.; Sokolov, A.; Meyerowitz, J.G.; Mathis, C.; et al. N-Myc Drives Neuroendocrine Prostate Cancer Initiated from Human Prostate Epithelial Cells. Cancer Cell 2016, 29, 536–547. [Google Scholar] [CrossRef] [Green Version]
- Qi, J.; Nakayama, K.; Cardiff, R.D.; Borowsky, A.D.; Kaul, K.; Williams, R.; Krajewski, S.; Mercola, D.; Carpenter, P.M.; Bowtell, D.; et al. Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors. Cancer Cell 2010, 18, 23–38. [Google Scholar] [CrossRef] [Green Version]
- Mosquera, J.M.; Beltran, H.; Park, K.; MacDonald, T.Y.; Robinson, B.D.; Tagawa, S.T.; Perner, S.; Bismar, T.A.; Erbersdobler, A.; Dhir, R.; et al. Concurrent AURKA and MYCN gene amplifications are harbingers of lethal treatment-related neuroendocrine prostate cancer. Neoplasia 2013, 15, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Bhagirath, D.; Yang, T.L.; Tabatabai, Z.L.; Majid, S.; Dahiya, R.; Tanaka, Y.; Saini, S. BRN4 Is a Novel Driver of Neuroendocrine Differentiation in Castration-Resistant Prostate Cancer and Is Selectively Released in Extracellular Vesicles with BRN2. Clin. Cancer Res. 2019, 25, 6532–6545. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.J.; Yun, E.J.; Lo, U.G.; Tai, Y.L.; Deng, S.; Hernandez, E.; Dang, A.; Chen, Y.A.; Saha, D.; Mu, P.; et al. The paracrine induction of prostate cancer progression by caveolin-1. Cell Death Dis. 2019, 10, 834. [Google Scholar] [CrossRef] [Green Version]
- Quaglia, F.; Krishn, S.R.; Daaboul, G.G.; Sarker, S.; Pippa, R.; Domingo-Domenech, J.; Kumar, G.; Fortina, P.; McCue, P.; Kelly, W.K.; et al. Small extracellular vesicles modulated by alphaVbeta3 integrin induce neuroendocrine differentiation in recipient cancer cells. J. Extracell. Vesicles 2020, 9, 1761072. [Google Scholar] [CrossRef]
- Dai, J.; Escara-Wilke, J.; Keller, J.M.; Jung, Y.; Taichman, R.S.; Pienta, K.J.; Keller, E.T. Primary prostate cancer educates bone stroma through exosomal pyruvate kinase M2 to promote bone metastasis. J. Exp. Med. 2019, 216, 2883–2899. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Selbach, M.; Schwanhausser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, 6478. [Google Scholar] [CrossRef]
- Title, A.C.; Hong, S.J.; Pires, N.D.; Hasenohrl, L.; Godbersen, S.; Stokar-Regenscheit, N.; Bartel, D.P.; Stoffel, M. Genetic dissection of the miR-200-Zeb1 axis reveals its importance in tumor differentiation and invasion. Nat. Commun. 2018, 9, 4671. [Google Scholar] [CrossRef]
- Lu, Y.X.; Yuan, L.; Xue, X.L.; Zhou, M.; Liu, Y.; Zhang, C.; Li, J.P.; Zheng, L.; Hong, M.; Li, X.N. Regulation of colorectal carcinoma stemness, growth, and metastasis by an miR-200c-Sox2-negative feedback loop mechanism. Clin. Cancer Res. 2014, 20, 2631–2642. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Feng, Z.; Zhang, X.; Lan, D.; Wu, Y. Up-regulation of microRNA-200c-3p inhibits invasion and migration of renal cell carcinoma cells via the SOX2-dependent Wnt/beta-catenin signaling pathway. Cancer Cell Int. 2019, 19, 231. [Google Scholar] [CrossRef] [Green Version]
- Beclin, C.; Follert, P.; Stappers, E.; Barral, S.; Core, N.; de Chevigny, A.; Magnone, V.; Lebrigand, K.; Bissels, U.; Huylebroeck, D.; et al. MiR-200 family controls late steps of postnatal forebrain neurogenesis via Zeb2 inhibition. Sci. Rep. 2016, 6, 35729. [Google Scholar] [CrossRef] [Green Version]
- Nandana, S.; Tripathi, M.; Duan, P.; Chu, C.Y.; Mishra, R.; Liu, C.; Jin, R.; Yamashita, H.; Zayzafoon, M.; Bhowmick, N.A.; et al. Bone Metastasis of Prostate Cancer Can Be Therapeutically Targeted at the TBX2-WNT Signaling Axis. Cancer Res. 2017, 77, 1331–1344. [Google Scholar] [CrossRef] [Green Version]
- Patel, G.K.; Khan, M.A.; Bhardwaj, A.; Srivastava, S.K.; Zubair, H.; Patton, M.C.; Singh, S.; Khushman, M.; Singh, A.P. Exosomes confer chemoresistance to pancreatic cancer cells by promoting ROS detoxification and miR-155-mediated suppression of key gemcitabine-metabolising enzyme, DCK. Br. J. Cancer 2017, 116, 609–619. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Wang, X. MiRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. MiRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef]
- Sharma, M.; Molehin, D.; Castro-Piedras, I.; Martinez, E.G.; Pruitt, K. Acetylation of conserved DVL-1 lysines regulates its nuclear translocation and binding to gene promoters in triple-negative breast cancer. Sci. Rep. 2019, 9, 16257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, S.; Yu, X. Bioinformatics analyses of publicly available NEPCa datasets. Am. J. Clin. Exp. Urol. 2019, 7, 327–340. [Google Scholar]
- Muralidharan-Chari, V.; Clancy, J.; Plou, C.; Romao, M.; Chavrier, P.; Raposo, G.; D’Souza-Schorey, C. ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr. Biol. 2009, 19, 1875–1885. [Google Scholar] [CrossRef] [Green Version]
- Patton, M.C.; Zubair, H.; Khan, M.A.; Singh, S.; Singh, A.P. Hypoxia alters the release and size distribution of extracellular vesicles in pancreatic cancer cells to support their adaptive survival. J. Cell Biochem. 2020, 121, 828–839. [Google Scholar] [CrossRef]
- Xia, L.; Han, Q.; Chi, C.; Zhu, Y.; Pan, J.; Dong, B.; Huang, Y.; Xia, W.; Xue, W.; Sha, J. Transcriptional regulation of PRKAR2B by miR-200b-3p/200c-3p and XBP1 in human prostate cancer. Biomed. Pharmacother. 2020, 124, 109863. [Google Scholar] [CrossRef]
- Liu, Y.N.; Yin, J.J.; Abou-Kheir, W.; Hynes, P.G.; Casey, O.M.; Fang, L.; Yi, M.; Stephens, R.M.; Seng, V.; Sheppard-Tillman, H.; et al. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene 2013, 32, 296–306. [Google Scholar] [CrossRef] [Green Version]
- Lynch, S.M.; O’Neill, K.M.; McKenna, M.M.; Walsh, C.P.; McKenna, D.J. Regulation of miR-200c and miR-141 by Methylation in Prostate Cancer. Prostate 2016, 76, 1146–1159. [Google Scholar] [CrossRef]
- Tseng, J.H.; Bisogna, M.; Hoang, L.N.; Olvera, N.; Rodriguez-Aguayo, C.; Lopez-Berestein, G.; Sood, A.K.; Levine, D.A.; Jelinic, P. MiR-200c-driven Mesenchymal-To-Epithelial Transition is a Therapeutic Target in Uterine Carcinosarcomas. Sci. Rep. 2017, 7, 3614. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Tian, W.; He, H.; Chen, F.; Huang, J.; Wang, X.; Chen, Z. Downregulation of miR200c3p contributes to the resistance of breast cancer cells to paclitaxel by targeting SOX2. Oncol. Rep. 2018, 40, 3821–3829. [Google Scholar]
- Carreira, S.; Dexter, T.J.; Yavuzer, U.; Easty, D.J.; Goding, C.R. Brachyury-related transcription factor Tbx2 and repression of the melanocyte-specific TRP-1 promoter. Mol. Cell. Biol. 1998, 18, 5099–5108. [Google Scholar] [CrossRef] [Green Version]
- Conlon, F.L.; Fairclough, L.; Price, B.M.; Casey, E.S.; Smith, J.C. Determinants of T box protein specificity. Development 2001, 128, 3749–3758. [Google Scholar] [CrossRef]
- Abrahams, A.; Parker, M.I.; Prince, S. The T-box transcription factor Tbx2: Its role in development and possible implication in cancer. IUBMB Life 2010, 62, 92–102. [Google Scholar] [CrossRef]
- Prince, S.; Carreira, S.; Vance, K.W.; Abrahams, A.; Goding, C.R. Tbx2 directly represses the expression of the p21(WAF1) cyclin-dependent kinase inhibitor. Cancer Res. 2004, 64, 1669–1674. [Google Scholar] [CrossRef] [Green Version]
- Kregel, S.; Kiriluk, K.J.; Rosen, A.M.; Cai, Y.; Reyes, E.E.; Otto, K.B.; Tom, W.; Paner, G.P.; Szmulewitz, R.Z.; Vander Griend, D.J. Sox2 is an androgen receptor-repressed gene that promotes castration-resistant prostate cancer. PLoS ONE 2013, 8, e53701. [Google Scholar] [CrossRef] [Green Version]
- Jin, R.J.; Wang, Y.; Masumori, N.; Ishii, K.; Tsukamoto, T.; Shappell, S.B.; Hayward, S.W.; Kasper, S.; Matusik, R.J. NE-10 neuroendocrine cancer promotes the LNCaP xenograft growth in castrated mice. Cancer Res. 2004, 64, 5489–5495. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zhang, H.; Qin, Y.; Chen, C.; Yang, J.; Song, N.; Gu, M. MicroRNA-200c-3p/ZEB2 loop plays a crucial role in the tumor progression of prostate carcinoma. Ann. Transl. Med. 2019, 7, 141. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhou, C.; Li, X.; Barnes, S.D.; Deng, S.; Hoover, E.; Chen, C.C.; Lee, Y.S.; Zhang, Y.; Wang, C.; et al. Loss of CHD1 Promotes Heterogeneous Mechanisms of Resistance to AR-Targeted Therapy via Chromatin Dysregulation. Cancer Cell 2020, 37, 584–598. [Google Scholar] [CrossRef]
- Vance, K.W.; Carreira, S.; Brosch, G.; Goding, C.R. Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer Res. 2005, 65, 2260–2268. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patel, G.K.; Dutta, S.; Mahmud Syed, M.; Ramachandran, S.; Sharma, M.; Rajamanickam, V.; Ganapathy, V.; DeGraff, D.J.; Pruitt, K.; Tripathi, M.; et al. TBX2 Drives Neuroendocrine Prostate Cancer through Exosome-Mediated Repression of miR-200c-3p. Cancers 2021, 13, 5020. https://doi.org/10.3390/cancers13195020
Patel GK, Dutta S, Mahmud Syed M, Ramachandran S, Sharma M, Rajamanickam V, Ganapathy V, DeGraff DJ, Pruitt K, Tripathi M, et al. TBX2 Drives Neuroendocrine Prostate Cancer through Exosome-Mediated Repression of miR-200c-3p. Cancers. 2021; 13(19):5020. https://doi.org/10.3390/cancers13195020
Chicago/Turabian StylePatel, Girijesh Kumar, Sayanika Dutta, Mosharaf Mahmud Syed, Sabarish Ramachandran, Monica Sharma, Venkatesh Rajamanickam, Vadivel Ganapathy, David J. DeGraff, Kevin Pruitt, Manisha Tripathi, and et al. 2021. "TBX2 Drives Neuroendocrine Prostate Cancer through Exosome-Mediated Repression of miR-200c-3p" Cancers 13, no. 19: 5020. https://doi.org/10.3390/cancers13195020