Oncogenic Serine 45-Deleted β-Catenin Remains Susceptible to Wnt Stimulation and APC Regulation in Human Colonocytes
Abstract
:1. Introduction
2. Results
2.1. Phosphorylation Sites Important For Β-Catenin Destruction Are Mutated in a Subset of Colorectal Cancers
2.2. Generation of a Novel Anti-APC Ιgy Antibody
2.3. Β-Catδs45 Associates With a Locally-Applied Wnt-3a Ligand
2.4. Wnt3a Exposure or APC-Depletion Increases Level of Β-Catδs45 Protein
2.5. Wnt Signaling Is Activated in HCT116βm Cells Following APC-Depletion or Wnt3a Exposure
2.6. β-catΔS45 Increases Nuclear Localization Upon APC Loss, But Not Upon Wnt Exposure
2.7. β-catΔS45 Is Phosphorylated at the GSK-3β Sites and Is Susceptible to Proteasomal Degradation
2.8. APC Truncation But Not β-Catenin Mutation Results in Elevated Wnt Target Gene Expression in Human Colorectal Cancers
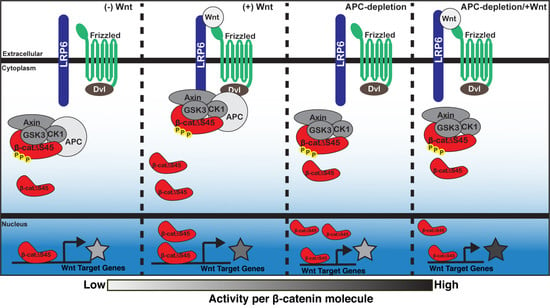
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Treatments
4.2. Analysis of CTNNB1 Mutation Frequency and mRNA Expression of Wnt Target Genes
4.3. CTNNB1 Sanger Sequencing and Alignment
4.4. Immobilization of Wnt Protein
4.5. Immunoblotting
4.6. Wnt-Bead Pull-Down
4.7. Generation of Anti-APC-M2 Chicken IgY Antibody
4.8. Luciferase Reporter Assay
4.9. Immunofluorescence and Analysis
4.10. Quantification and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Logan, C.Y.; Nusse, R. The Wnt Signaling Pathway in Development and Disease. Annu. Rev. Cell Dev. Biol. 2004, 20, 781–810. [Google Scholar] [CrossRef] [Green Version]
- Clevers, H. Wnt/β-Catenin Signaling in Development and Disease. Cell 2006, 127, 469–480. [Google Scholar] [CrossRef] [Green Version]
- MacDonald, B.T.; Tamai, K.; He, X. Wnt/β-Catenin Signaling: Components, Mechanisms, and Diseases. Dev. Cell 2009, 17, 9–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [CrossRef] [PubMed] [Green Version]
- Stamos, J.L.; Weis, W.I. The β-Catenin Destruction Complex. Cold Spring Harb. Perspect. Biol. 2013, 5, a007898. [Google Scholar] [CrossRef]
- Aberle, H.; Bauer, A.; Stappert, J.; Kispert, A.; Kemler, R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997, 16, 3797–3804. [Google Scholar] [CrossRef] [Green Version]
- Kitagawa, M.; Hatakeyama, S.; Shirane, M.; Matsumoto, M.; Ishida, N.; Hattori, K.; Nakamichi, I.; Kikuchi, A.; Nakayama, K.; Nakayama, K. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of beta-catenin. EMBO J. 1999, 18, 2401–2410. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Li, Y.; Semenov, M.; Han, C.; Baeg, G.-H.; Tan, Y.; Zhang, Z.; Lin, X.; He, X. Control of β-Catenin Phosphorylation/Degradation by a Dual-Kinase Mechanism. Cell 2002, 108, 837–847. [Google Scholar] [CrossRef] [Green Version]
- Bilić, J.; Huang, Y.-L.; Davidson, G.; Zimmermann, T.; Cruciat, C.-M.; Bienz, M.; Niehrs, C. Wnt Induces LRP6 Signalosomes and Promotes Dishevelled-Dependent LRP6 Phosphorylation. Science 2007, 316, 1619–1622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamai, K.; Semenov, M.; Kato, Y.; Spokony, R.; Liu, C.; Katsuyama, Y.; Hess, F.; Saint-Jeannet, J.-P.; He, X. LDL-receptor-related proteins in Wnt signal transduction. Nature 2000, 407, 530. [Google Scholar] [CrossRef]
- Amit, S.; Hatzubai, A.; Birman, Y.; Andersen, J.S.; Ben-Shushan, E.; Mann, M.; Ben-Neriah, Y.; Alkalay, I. Axin-mediated CKI phosphorylation of β-catenin at Ser 45: A molecular switch for the Wnt pathway. Genes Dev. 2002, 16, 1066–1076. [Google Scholar] [CrossRef] [Green Version]
- Fiol, C.J.; Mahrenholz, A.M.; Wang, Y.; Roeske, R.W.; Roach, P.J. Formation of protein kinase recognition sites by covalent modification of the substrate. Molecular mechanism for the synergistic action of casein kinase II and glycogen synthase kinase 3. J. Biol. Chem. 1987, 262, 14042–14048. [Google Scholar]
- Frame, S.; Cohen, P.; Biondi, R.M. A Common Phosphate Binding Site Explains the Unique Substrate Specificity of GSK3 and Its Inactivation by Phosphorylation. Mol. Cell 2001, 7, 1321–1327. [Google Scholar] [CrossRef]
- Hagen, T.; Vidal-Puig, A. Characterisation of the phosphorylation of β-catenin at the GSK-3 priming site Ser45. Biochem. Biophys. Res. Commun. 2002, 294, 324–328. [Google Scholar] [CrossRef]
- Hart, M.; Concordet, J.-P.; Lassot, I.; Albert, I.; del los Santos, R.; Durand, H.; Perret, C.; Rubinfeld, B.; Margottin, F.; Benarous, R.; et al. The F-box protein β-TrCP associates with phosphorylated β-catenin and regulates its activity in the cell. Curr. Biol. 1999, 9, 207–211. [Google Scholar] [CrossRef] [Green Version]
- Latres, E.; Chiaur, D.S.; Pagano, M. The human F box protein β-Trcp associates with the Cul1/Skp1 complex and regulates the stability of β-catenin. Oncogene 1999, 18, 849–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Kato, Y.; Zhang, Z.; Do, V.M.; Yankner, B.A.; He, X. β-Trcp couples β-catenin phosphorylation-degradation and regulates Xenopus axis formation. Proc. Natl. Acad. Sci. USA 1999, 96, 6273–6278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winston, J.T.; Strack, P.; Beer-Romero, P.; Chu, C.Y.; Elledge, S.J.; Harper, J.W. The SCFβ-TRCP–ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 1999, 13, 270–283. [Google Scholar] [CrossRef]
- Albuquerque, C.; Breukel, C.; van der Luijt, R.; Fidalgo, P.; Lage, P.; Slors, F.J.M.; Leitão, C.N.; Fodde, R.; Smits, R. The ‘just-right’ signaling model: APC somatic mutations are selected based on a specific level of activation of the β-catenin signaling cascade. Hum. Mol. Genet. 2002, 11, 1549–1560. [Google Scholar] [CrossRef] [Green Version]
- Neufeld, K.L.; Nix, D.A.; Bogerd, H.; Kang, Y.; Beckerle, M.C.; Cullen, B.R.; White, R.L. Adenomatous polyposis coli protein contains two nuclear export signals and shuttles between the nucleus and cytoplasm. Proc. Natl. Acad. Sci. USA 2000, 97, 12085–12090. [Google Scholar] [CrossRef] [Green Version]
- Neufeld, K.L.; Zhang, F.; Cullen, B.R.; White, R.L. APC-mediated downregulation of -catenin activity involves nuclear sequestration and nuclear export. EMBO Rep. 2000, 1, 519–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosin-Arbesfeld, R.; Townsley, F.; Bienz, M. The APC tumour suppressor has a nuclear export function. Nature 2000, 406, 1009. [Google Scholar] [CrossRef] [PubMed]
- Rosin-Arbesfeld, R.; Cliffe, A.; Brabletz, T.; Bienz, M. Nuclear export of the APC tumour suppressor controls β-catenin function in transcription. EMBO J. 2003, 22, 1101–1113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sierra, J.; Yoshida, T.; Joazeiro, C.A.; Jones, K.A. The APC tumor suppressor counteracts β-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 2006, 20, 586–600. [Google Scholar] [CrossRef] [Green Version]
- Li, V.S.W.; Ng, S.S.; Boersema, P.J.; Low, T.Y.; Karthaus, W.R.; Gerlach, J.P.; Mohammed, S.; Heck, A.J.R.; Maurice, M.M.; Mahmoudi, T.; et al. Wnt Signaling through Inhibition of β-Catenin Degradation in an Intact Axin1 Complex. Cell 2012, 149, 1245–1256. [Google Scholar] [CrossRef] [Green Version]
- Parker, T.W.; Neufeld, K.L. APC controls Wnt-induced β-catenin destruction complex recruitment in human colonocytes. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.; Fu, C.; Ishikawa, S.; Stella, A.; Kojima, M.; Shitoh, K.; Schreiber, E.M.; Day, B.W.; Liu, B. APC Is Essential for Targeting Phosphorylated β-Catenin to the SCFβ-TrCP Ubiquitin Ligase. Mol. Cell 2008, 32, 652–661. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, W.; Evans, P.M.; Chen, X.; He, X.; Liu, C. Adenomatous Polyposis Coli (APC) Differentially Regulates β-Catenin Phosphorylation and Ubiquitination in Colon Cancer Cells. J. Biol. Chem. 2006, 281, 17751–17757. [Google Scholar] [CrossRef] [Green Version]
- Morin, P.J.; Sparks, A.B.; Korinek, V.; Barker, N.; Clevers, H.; Vogelstein, B.; Kinzler, K.W. Activation of β-Catenin-Tcf Signaling in Colon Cancer by Mutations in β-Catenin or APC. Science 1997, 275, 1787–1790. [Google Scholar] [CrossRef] [Green Version]
- Rebouissou, S.; Franconi, A.; Calderaro, J.; Letouzé, E.; Imbeaud, S.; Pilati, C.; Nault, J.-C.; Couchy, G.; Laurent, A.; Balabaud, C.; et al. Genotype-phenotype correlation of CTNNB1 mutations reveals different ß-catenin activity associated with liver tumor progression. Hepatology 2016, 64, 2047–2061. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Vogelstein, B.; Kinzler, K.W. Phosphorylation of β-Catenin at S33, S37, or T41 Can Occur in the Absence of Phosphorylation at T45 in Colon Cancer Cells. Cancer Res. 2003, 63, 5234–5235. [Google Scholar] [PubMed]
- Chan, T.A.; Wang, Z.; Dang, L.H.; Vogelstein, B.; Kinzler, K.W. Targeted inactivation of CTNNB1 reveals unexpected effects of β-catenin mutation. Proc. Natl. Acad. Sci. USA 2002, 99, 8265–8270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albuquerque, C.; Bakker, E.R.M.; van Veelen, W.; Smits, R. Colorectal cancers choosing sides. Biochim. Biophys. Acta BBA Rev. Cancer 2011, 1816, 219–231. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Jeong, S. Mutation Hotspots in the β-Catenin Gene: Lessons from the Human Cancer Genome Databases. Mol. Cells 2019, 42, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Giannakis, M.; Mu, X.J.; Shukla, S.A.; Qian, Z.R.; Cohen, O.; Nishihara, R.; Bahl, S.; Cao, Y.; Amin-Mansour, A.; Yamauchi, M.; et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016, 15, 857–865. [Google Scholar] [CrossRef] [Green Version]
- Seshagiri, S.; Stawiski, E.W.; Durinck, S.; Modrusan, Z.; Storm, E.E.; Conboy, C.B.; Chaudhuri, S.; Guan, Y.; Janakiraman, V.; Jaiswal, B.S.; et al. Recurrent R-spondin fusions in colon cancer. Nature 2012, 488, 660–664. [Google Scholar] [CrossRef]
- Yaeger, R.; Chatila, W.K.; Lipsyc, M.D.; Hechtman, J.F.; Cercek, A.; Sanchez-Vega, F.; Jayakumaran, G.; Middha, S.; Zehir, A.; Donoghue, M.T.A.; et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell 2018, 33, 125–136.e3. [Google Scholar] [CrossRef]
- Gao, C.; Wang, Y.; Broaddus, R.; Sun, L.; Xue, F.; Zhang, W. Exon 3 mutations of CTNNB1 drive tumorigenesis: A review. Oncotarget 2017, 9, 5492–5508. [Google Scholar] [CrossRef] [Green Version]
- Spink, K.E.; Fridman, S.G.; Weis, W.I. Molecular mechanisms of β-catenin recognition by adenomatous polyposis coli revealed by the structure of an APC–β-catenin complex. EMBO J. 2001, 20, 6203–6212. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Clements, W.K.; Kimelman, D.; Xu, W. Crystal structure of a β-catenin/Axin complex suggests a mechanism for the β-catenin destruction complex. Genes Dev. 2003, 17, 2753–2764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prieve, M.G.; Waterman, M.L. Nuclear Localization and Formation of β-Catenin–Lymphoid Enhancer Factor 1 Complexes Are Not Sufficient for Activation of Gene Expression. Mol. Cell. Biol. 1999, 19, 4503–4515. [Google Scholar] [CrossRef] [Green Version]
- Brocardo, M.; Näthke, I.S.; Henderson, B.R. Redefining the subcellular location and transport of APC: New insights using a panel of antibodies. EMBO Rep. 2005, 6, 184–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, M.L.; Roberts, G.T.; Stuart, N.; Wakeman, J.A. Analysis of a panel of antibodies to APC reveals consistent activity towards an unidentified protein. Br. J. Cancer 2007, 97, 384–390. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Azuma, Y.; Friedman, D.B.; Coffey, R.J.; Neufeld, K.L. Novel association of APC with intermediate filaments identified using a new versatile APC antibody. BMC Cell Biol. 2009, 10, 75. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, Y.; Hayashi, S.; Levine, A.; Wieschaus, E. Regulation of Armadillo by a Drosophila APC Inhibits Neuronal Apoptosis during Retinal Development. Cell 1998, 93, 1171–1182. [Google Scholar] [CrossRef] [Green Version]
- Sansom, O.J.; Reed, K.R.; Hayes, A.J.; Ireland, H.; Brinkmann, H.; Newton, I.P.; Batlle, E.; Simon-Assmann, P.; Clevers, H.; Nathke, I.S.; et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 2004, 18, 1385–1390. [Google Scholar] [CrossRef] [Green Version]
- Bottomly, D.; Kyler, S.L.; McWeeney, S.K.; Yochum, G.S. Identification of β-catenin binding regions in colon cancer cells using ChIP-Seq. Nucleic Acids Res. 2010, 38, 5735–5745. [Google Scholar] [CrossRef] [Green Version]
- Hatzis, P.; van der Flier, L.G.; van Driel, M.A.; Guryev, V.; Nielsen, F.; Denissov, S.; Nijman, I.J.; Koster, J.; Santo, E.E.; Welboren, W.; et al. Genome-Wide Pattern of TCF7L2/TCF4 Chromatin Occupancy in Colorectal Cancer Cells. Mol. Cell. Biol. 2008, 28, 2732–2744. [Google Scholar] [CrossRef] [Green Version]
- Sutherland, C. What Are the Bona Fide GSK3 Substrates? Available online: https://www.hindawi.com/journals/ijad/2011/505607/ (accessed on 21 April 2020).
- Olmeda, D.; Castel, S.; Vilaró, S.; Cano, A. β-Catenin Regulation during the Cell Cycle: Implications in G2/M and Apoptosis. Mol. Biol. Cell 2003, 14, 2844–2860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito-Diaz, K.; Benchabane, H.; Tiwari, A.; Tian, A.; Li, B.; Thompson, J.J.; Hyde, A.S.; Sawyer, L.M.; Jodoin, J.N.; Santos, E.; et al. APC Inhibits Ligand-Independent Wnt Signaling by the Clathrin Endocytic Pathway. Dev. Cell 2018, 44, 566–581.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Habib, S.J.; Chen, B.-C.; Tsai, F.-C.; Anastassiadis, K.; Meyer, T.; Betzig, E.; Nusse, R. A Localized Wnt Signal Orients Asymmetric Stem Cell Division in Vitro. Science 2013, 339, 1445–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korinek, V.; Barker, N.; Morin, P.J.; van Wichen, D.; de Weger, R.; Kinzler, K.W.; Vogelstein, B.; Clevers, H. Constitutive Transcriptional Activation by a β-Catenin-Tcf Complex in APC−/− Colon Carcinoma. Science 1997, 275, 1784–1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McQuin, C.; Goodman, A.; Chernyshev, V.; Kamentsky, L.; Cimini, B.A.; Karhohs, K.W.; Doan, M.; Ding, L.; Rafelski, S.M.; Thirstrup, D.; et al. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 2018, 16, e2005970. [Google Scholar] [CrossRef] [Green Version]
- CellProfiler/Tutorials. Available online: https://github.com/CellProfiler/tutorials (accessed on 29 July 2020).






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parker, T.W.; Rudeen, A.J.; Neufeld, K.L. Oncogenic Serine 45-Deleted β-Catenin Remains Susceptible to Wnt Stimulation and APC Regulation in Human Colonocytes. Cancers 2020, 12, 2114. https://doi.org/10.3390/cancers12082114
Parker TW, Rudeen AJ, Neufeld KL. Oncogenic Serine 45-Deleted β-Catenin Remains Susceptible to Wnt Stimulation and APC Regulation in Human Colonocytes. Cancers. 2020; 12(8):2114. https://doi.org/10.3390/cancers12082114
Chicago/Turabian StyleParker, Taybor W., Aaron J. Rudeen, and Kristi L. Neufeld. 2020. "Oncogenic Serine 45-Deleted β-Catenin Remains Susceptible to Wnt Stimulation and APC Regulation in Human Colonocytes" Cancers 12, no. 8: 2114. https://doi.org/10.3390/cancers12082114