Pilot Multi-Omic Analysis of Human Bile from Benign and Malignant Biliary Strictures: A Machine-Learning Approach
Abstract
:1. Introduction
2. Results
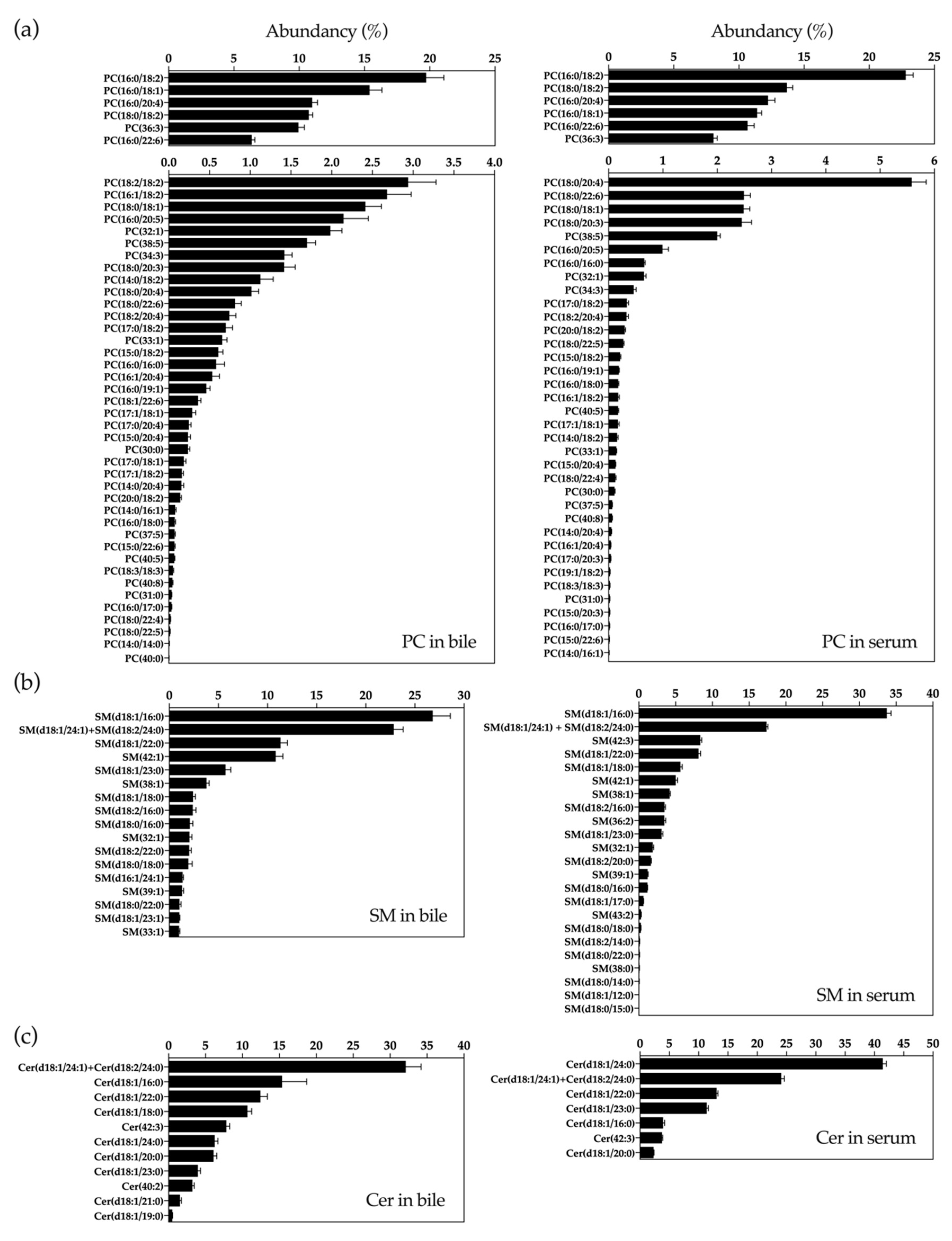
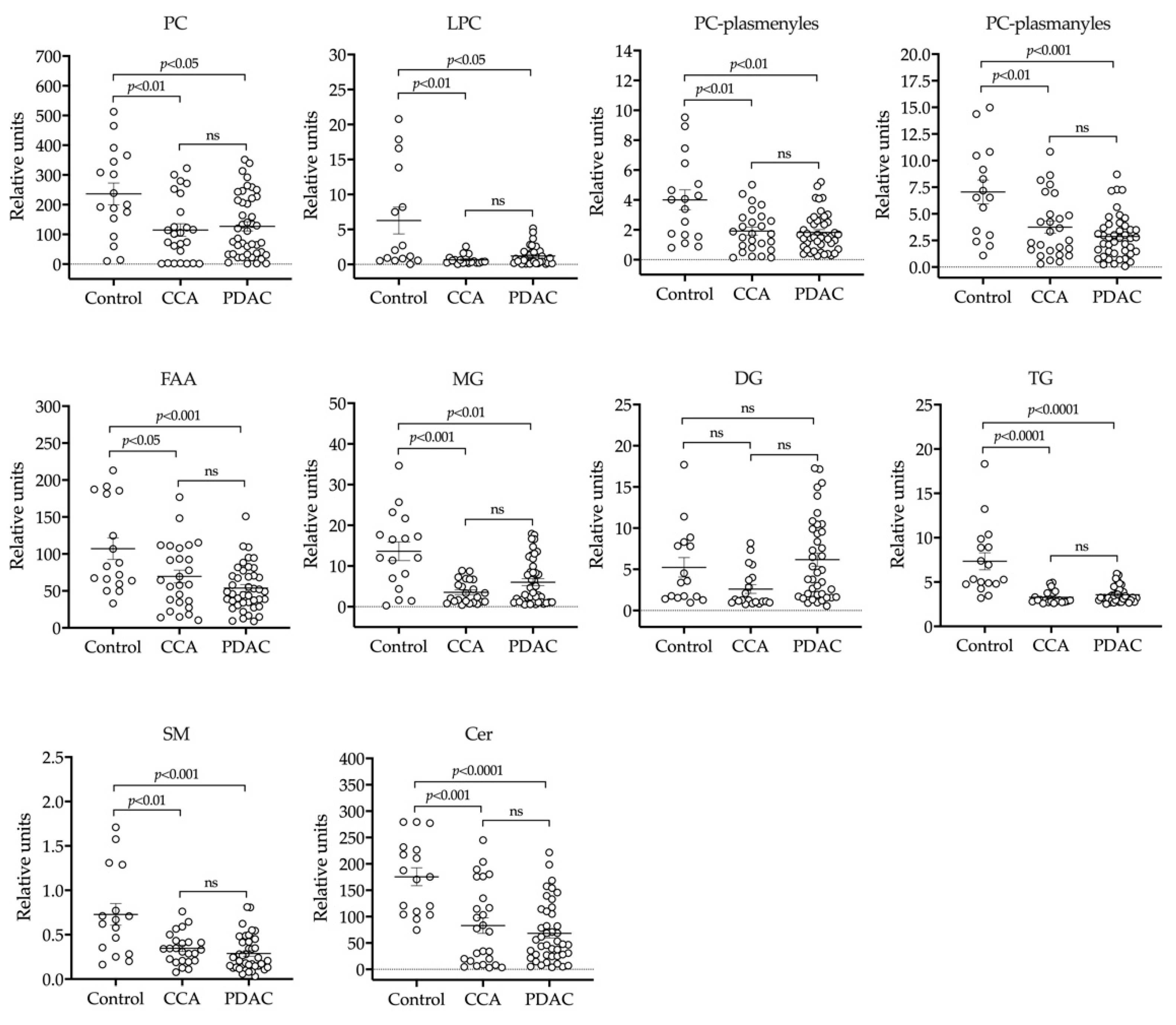
2.1. UHPLC-MS Lipidomic Analysis of Bile
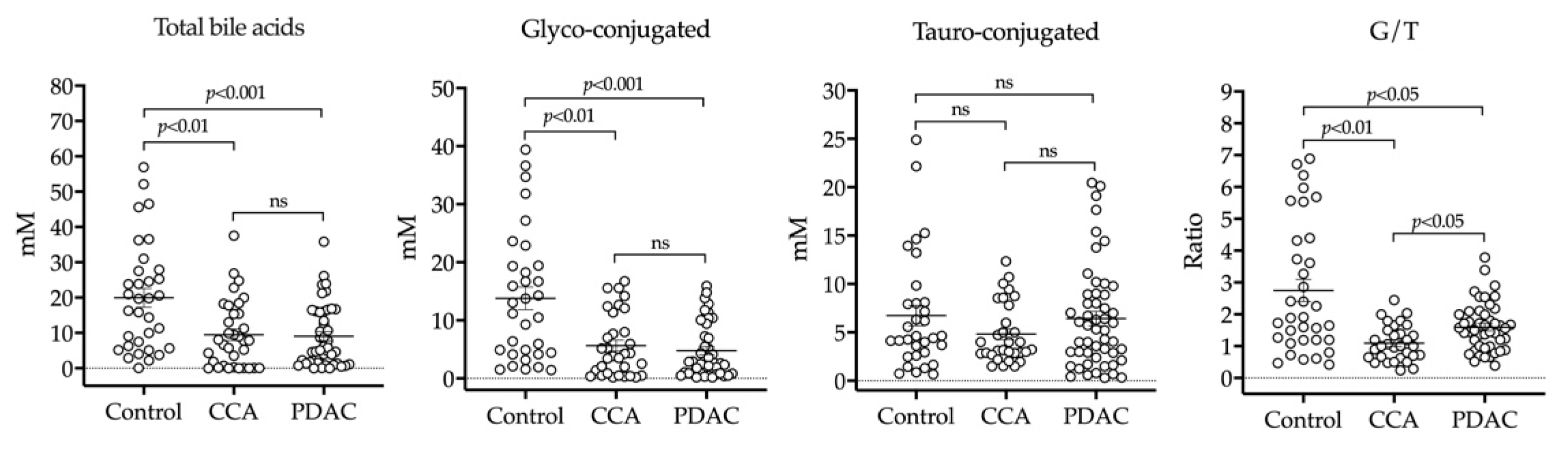
2.2. HPLC-MS/MS Analysis of BAs in Bile
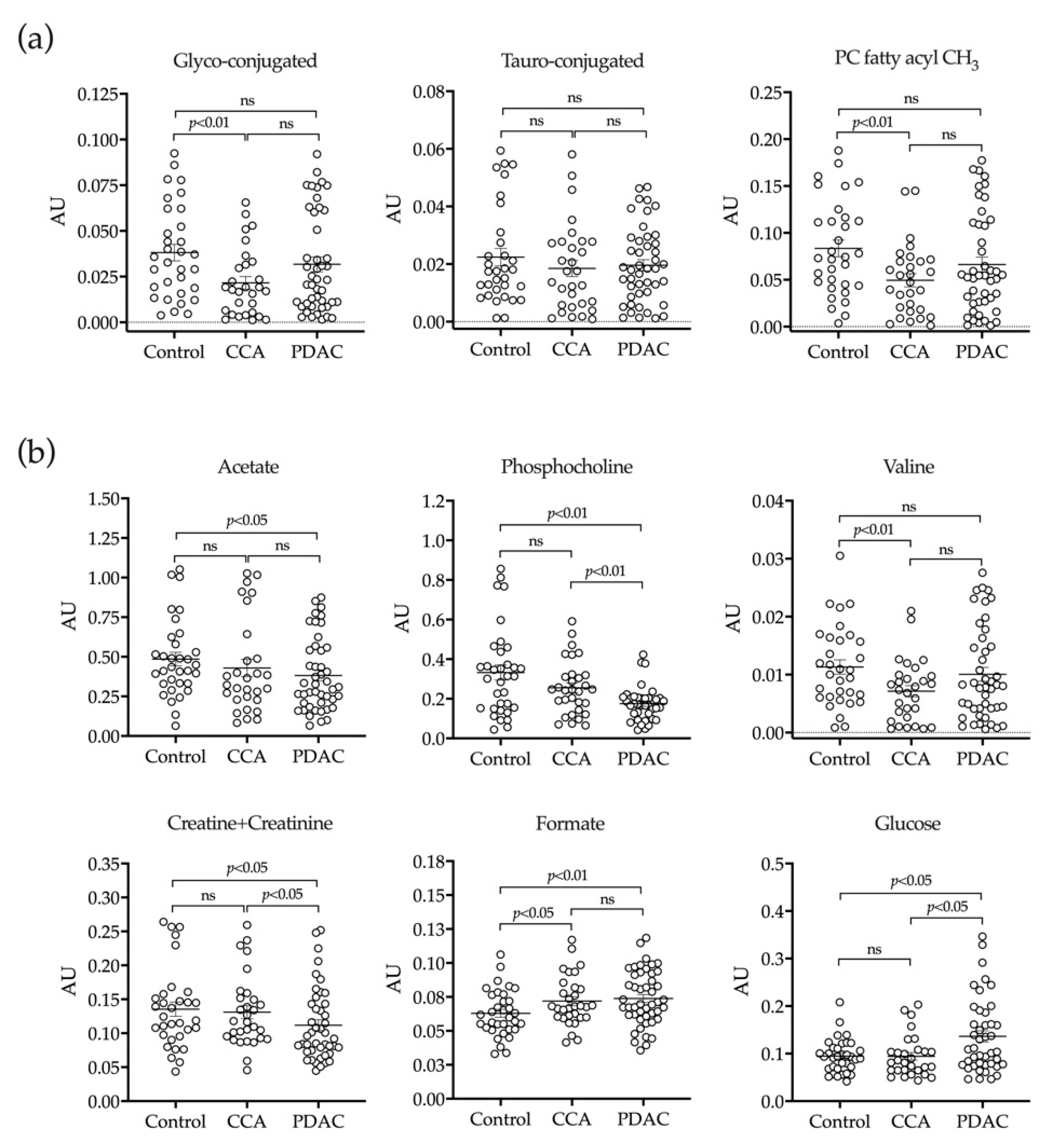
2.3. H-NMR Analysis of Bile
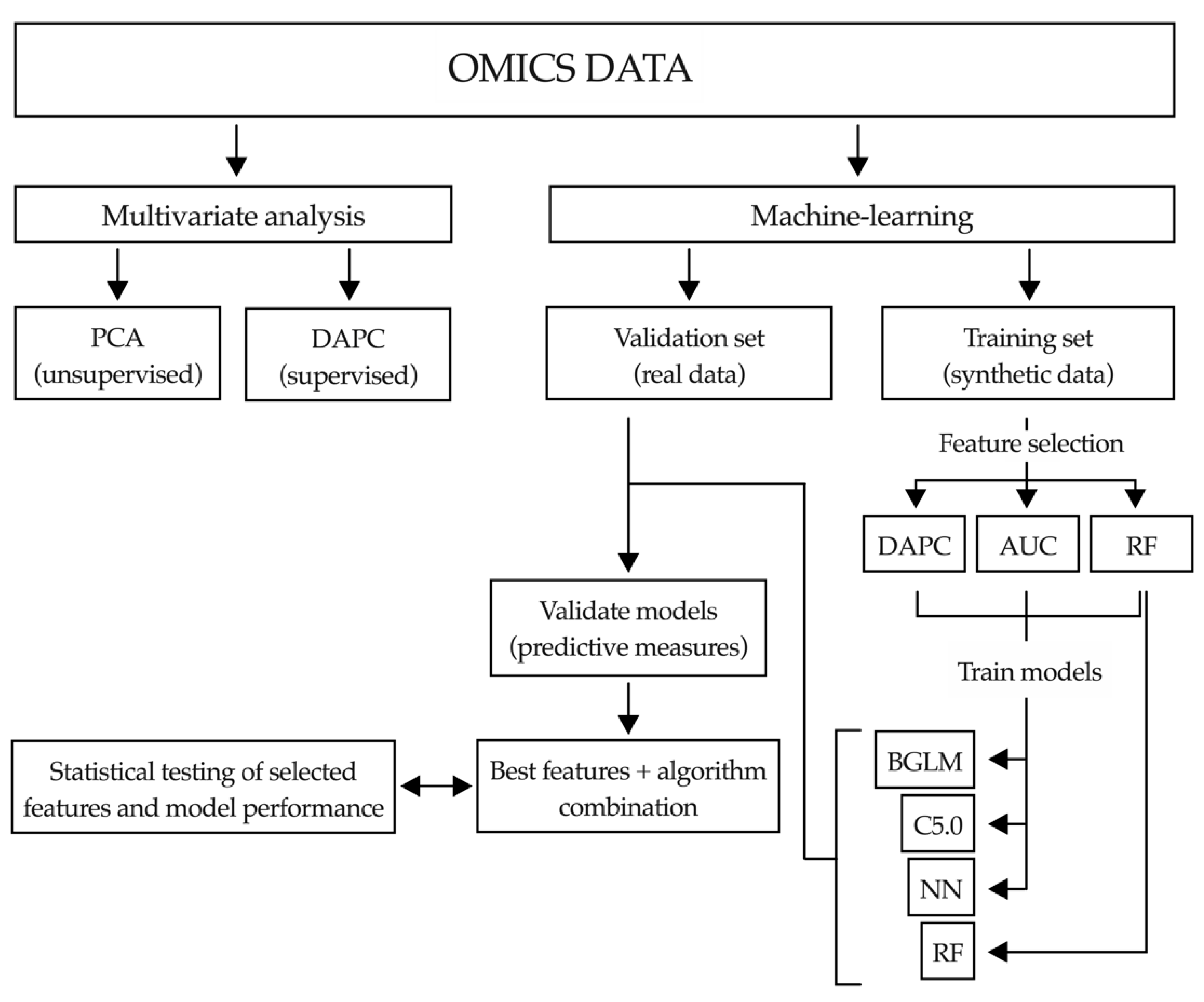
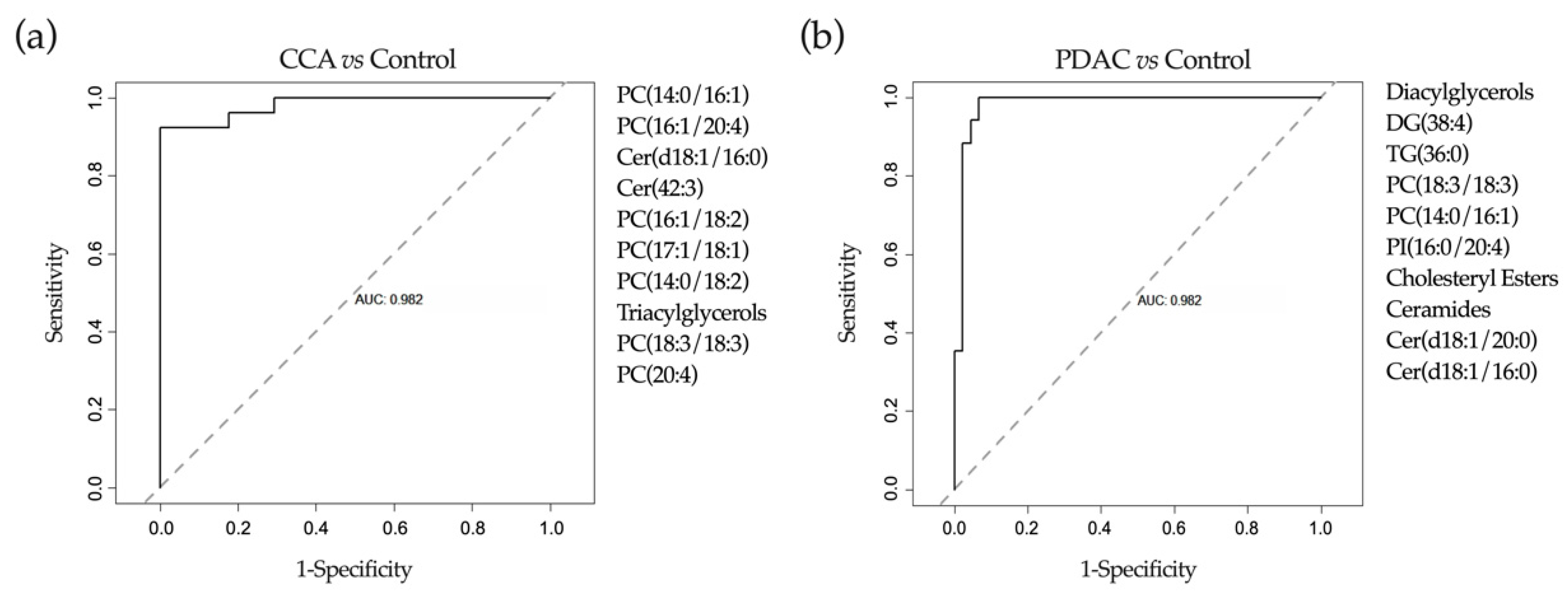
2.4. Application of Machine-Learning Methods to Metabolomic Data to Differentiate between Benign and Malignant Biliary Stenoses
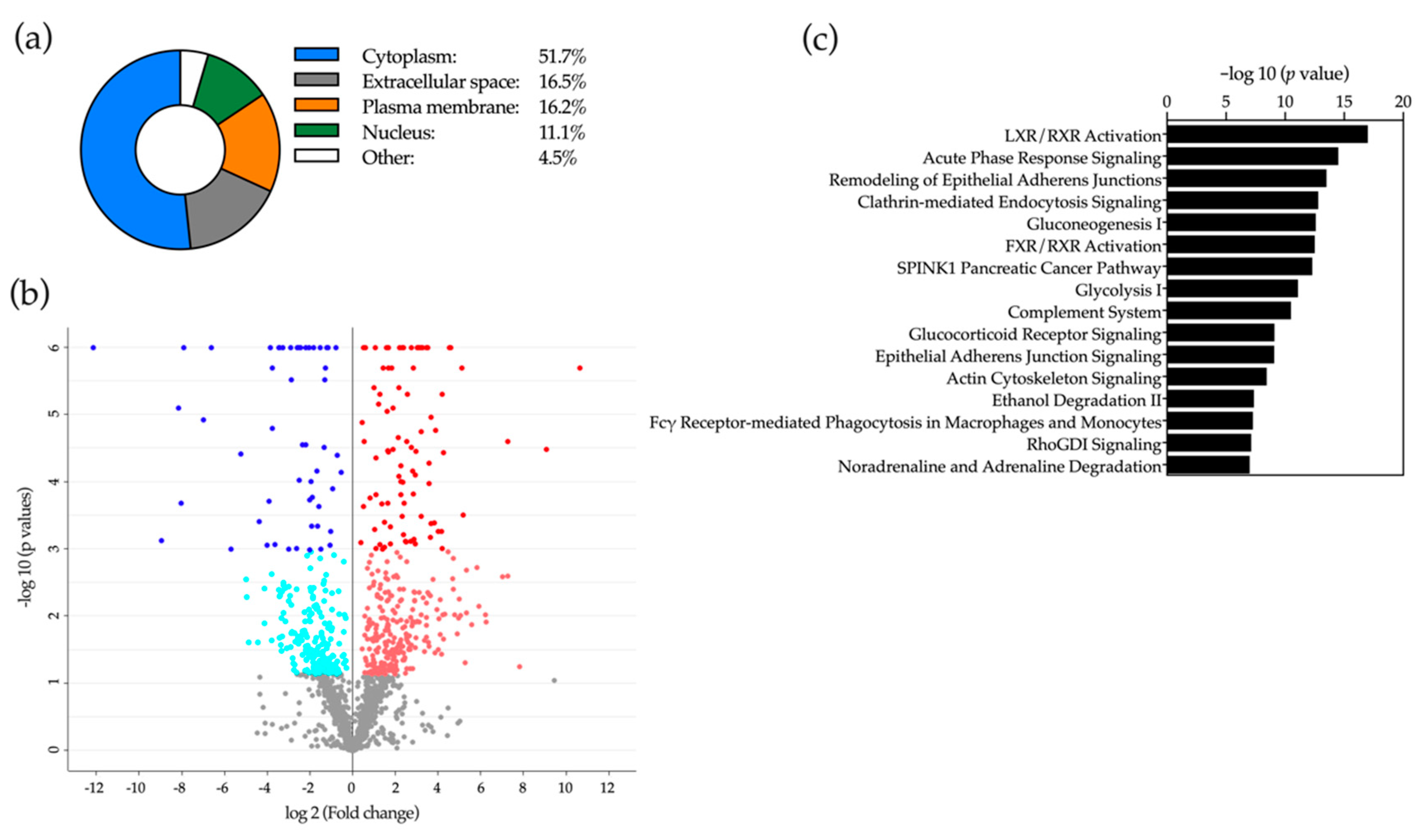
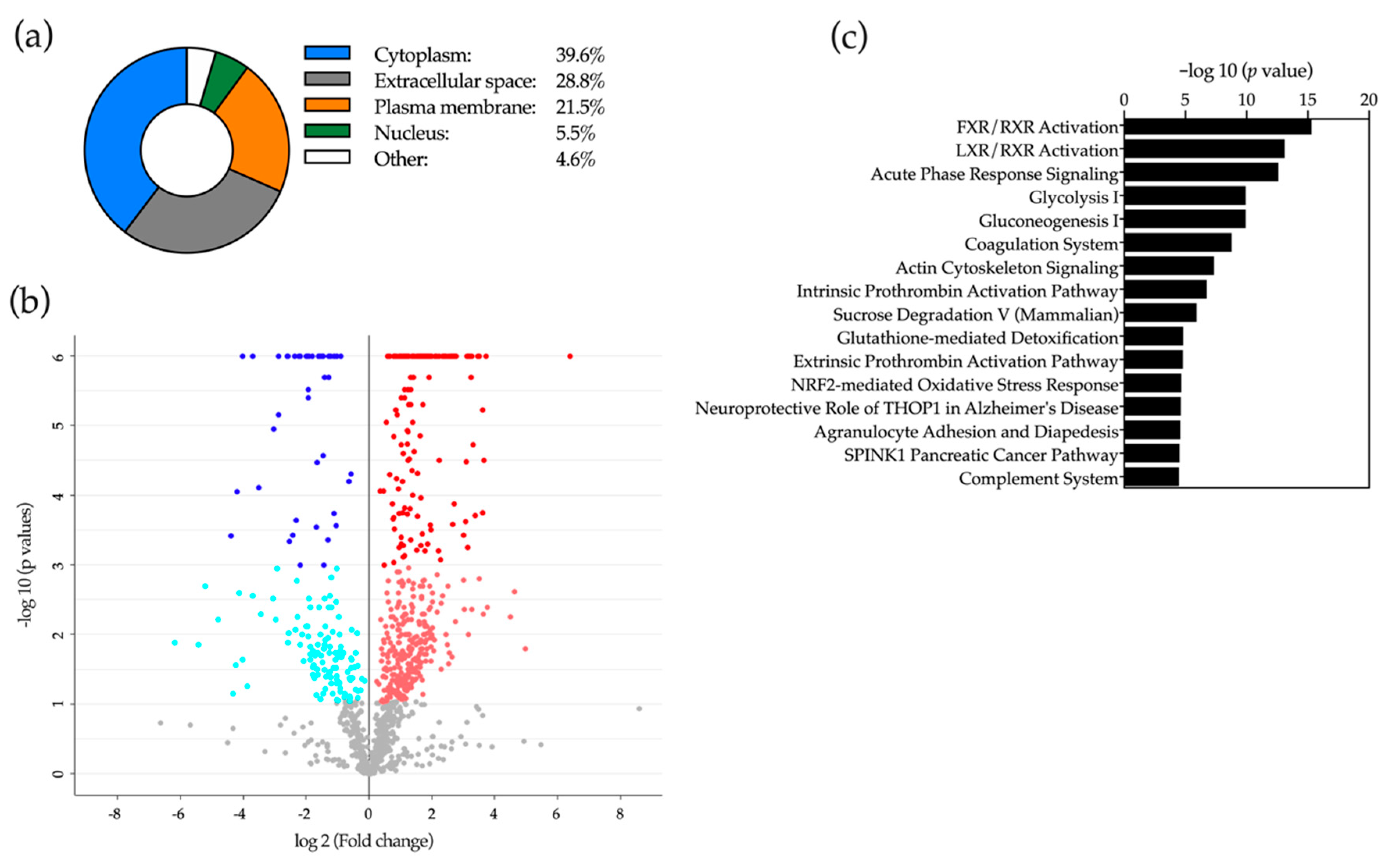
2.5. Proteomic Analysis of Bile
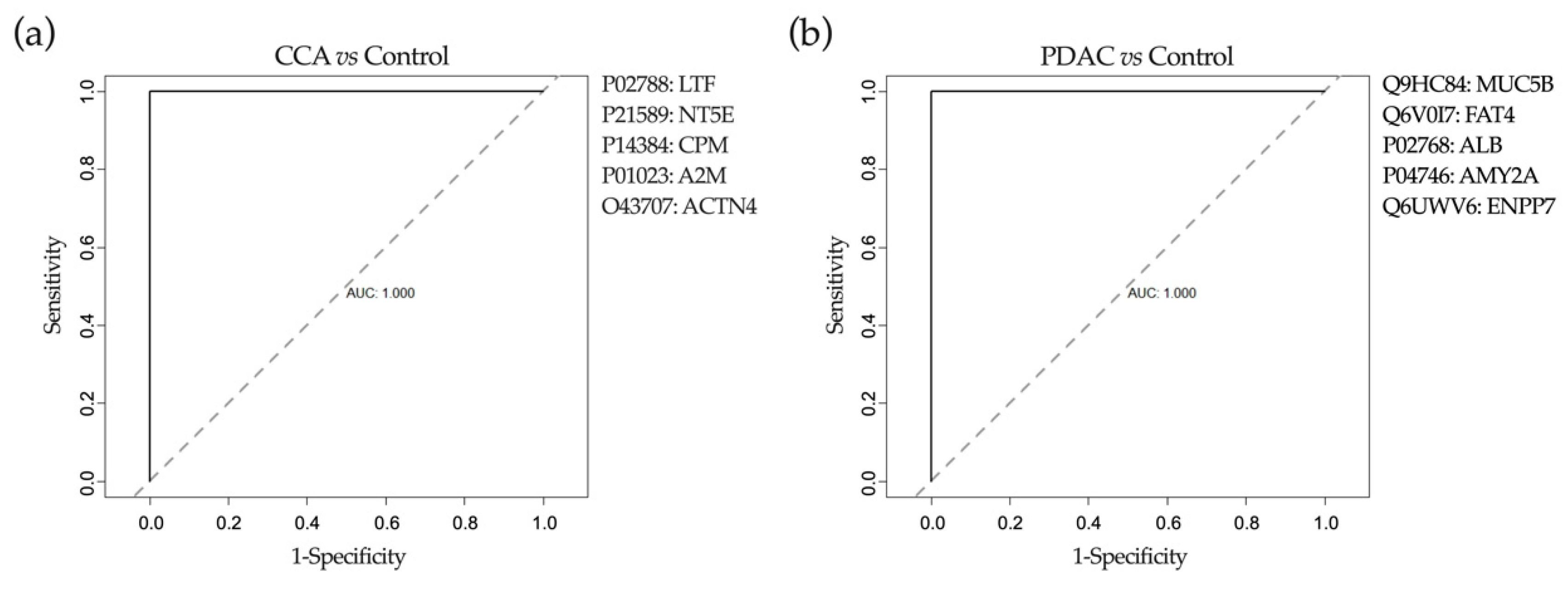
2.6. Application of Machine-Learning Methods to Bile Proteomic Data to Differentiate between Benign and Malignant Stenoses
3. Discussion
4. Materials and Methods
4.1. Patient Population and Samples Collection
4.2. Lipidomic Analyses
4.2.1. Lipid Extraction and Uhplc-Ms Analysis
4.2.2. Lipidomics Data Analysis
4.3. Analysis of BAs
4.4. H-NMR Analysis
4.4.1. Sample Preparation
4.4.2. H-NMR Experiments and Metabolite Quantification
4.5. Proteomic Analyses
4.5.1. Sample Preparation
4.5.2. LC-MS Analysis
4.5.3. Data Analysis and Quantification
4.6. Data Analysis and Machine Learning
4.6.1. Descriptive and Inferential Statistics
4.6.2. Machine-Learning Pipeline
Multivariate Analysis
Data Imputation
Synthetic Data Generation
Feature Selection
Artificial Intelligence Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hofmann, A.F. Bile composition. In Encyclopedia of Gastroenterology; Johnson, L.R., Ed.; Academic Press: Amsterdam, The Netherlands, 2004; pp. 176–184. [Google Scholar]
- Esteller, A. Physiology of bile secretion. World J. Gastroenterol. 2008, 14, 5641–5649. [Google Scholar] [CrossRef]
- Farina, A.; Delhaye, M.; Lescuyer, P.; Dumonceau, J.M. Bile proteome in health and disease. Compr. Physiol. 2014, 4, 91–108. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A. Human bile as a rich source of biomarkers for hepatopancreatobiliary cancers. Biomark. Med. 2010, 4, 299–314. [Google Scholar] [CrossRef]
- Hegyi, P.; Maléth, J.; Walters, J.R.; Hofmann, A.F.; Keely, S.J. Guts and gall: Bile acids in regulation of intestinal epithelial function in health and disease. Physiol. Rev. 2018, 98, 1983–2023. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Song, X.; Khan, S.; Li, Y.; Guo, Z.; Li, C.; Wang, S.; Dong, W.; Liu, W.; Wang, B.; et al. The gut microbiota at the intersection of bile acids and intestinal carcinogenesis: An old story, yet mesmerizing. Int. J. Cancer 2020, 146, 1780–1790. [Google Scholar] [CrossRef]
- Kummen, M.; Hov, J.R. The gut microbial influence on cholestatic liver disease. Liver Int. 2019, 39, 1186–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagana Gowda, G.A. NMR spectroscopy for discovery and quantitation of biomarkers of disease in human bile. Bioanalysis 2011, 3, 1877–1890. [Google Scholar] [CrossRef] [PubMed]
- Lourdusamy, V.; Tharian, B.; Navaneethan, U. Biomarkers in bile-complementing advanced endoscopic imaging in the diagnosis of indeterminate biliary strictures. World J. Gastrointest. Endosc. 2015, 7, 308. [Google Scholar] [CrossRef] [PubMed]
- Ijare, O.B.; Bezabeh, T.; Albiin, N.; Bergquist, A.; Arnelo, U.; Lindberg, B.; Smith, I.C.P. Simultaneous quantification of glycine- and taurine-conjugated bile acids, total bile acids, and choline-containing phospholipids in human bile using 1H NMR spectroscopy. J. Pharm. Biomed. Anal. 2010, 53, 667–673. [Google Scholar] [CrossRef] [Green Version]
- Bala, L.; Tripathi, P.; Bhatt, G.; Das, K.; Roy, R.; Choudhuri, G.; Khetrapal, C.L. 1H and 31P NMR studies indicate reduced bile constituents in patients with biliary obstruction and infection. NMR Biomed. 2009, 22, 220–228. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A.; Shanaiah, N.; Cooper, A.; Maluccio, M.; Raftery, D. Bile acids conjugation in human bile is not random: New insights from 1H-NMR spectroscopy at 800 MHz. Lipids 2009, 44, 527–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharif, A.W.; Williams, H.R.T.; Lampejo, T.; Khan, S.A.; Bansi, D.S.; Westaby, D.; Thillainayagam, A.V.; Thomas, H.C.; Cox, I.J.; Taylor-Robinson, S.D. Metabolic profiling of bile in cholangiocarcinoma using in vitro magnetic resonance spectroscopy. HPB 2010, 12, 396–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hay, D.W.; Carey, M.C. Chemical species of lipids in bile. Hepatology 1990, 12, 6S–14S. [Google Scholar] [PubMed]
- Gauss, A.; Ehehalt, R.; Lehmann, W.D.; Erben, G.; Weiss, K.H.; Schaefer, Y.; Kloeters-Plachky, P.; Stiehl, A.; Stremmel, W.; Sauer, P.; et al. Biliary phosphatidylcholine and lysophosphatidylcholine profiles in sclerosing cholangitis. World J. Gastroenterol. 2013, 19, 5454–5463. [Google Scholar] [CrossRef]
- Alvaro, D.; Cantafora, A.; Attili, A.F.; Ginanni Corradini, S.; De Luca, C.; Minervini, G.; Di Blase, A.; Angelico, M. Relationships between bile salts hydrophilicity and phospholipid composition in bile of various animal species. Comp. Biochem. Physiol. Part B Biochem. 1986, 83, 551–554. [Google Scholar] [CrossRef]
- Nibbering, C.P.; Carey, M.C. Sphingomyelins of rat liver: Biliary enrichment with molecular species containing 16:0 fatty acids as compared to canalicular-enriched plasma membranes. J. Membr. Biol. 1999, 167, 165–171. [Google Scholar] [CrossRef]
- Moschetta, A.; VanBerge-Henegouwen, G.P.; Portincasa, P.; Palasciano, G.; Groen, A.K.; Van Erpecum, K.J. Sphingomyelin exhibits greatly enhanced protection compared with egg yolk phosphatidylcholine against detergent bile salts. J. Lipid Res. 2000, 41, 916–924. [Google Scholar]
- Albiin, N.; Smith, I.C.P.; Arnelo, U.; Lindberg, B.; Bergquist, A.; Dolenko, B.; Bryksina, N.; Bezabeh, T. Detection of cholangiocarcinoma with magnetic resonance spectroscopy of bile in patients with and without primary sclerosing cholangitis. Acta Radiol. 2008, 49, 855–862. [Google Scholar] [CrossRef]
- Van Erpecum, K.J. Pathogenesis of cholesterol and pigment gallstones: An update. Clin. Res. Hepatol. Gastroenterol. 2011, 35, 281–287. [Google Scholar] [CrossRef]
- Zakarias, T.; Bunkenborg, J.; Gronborg, M.; Molina, H.; Thuluvath, P.J.; Argani, P.; Goggins, M.G.; Maitra, A.; Pandey, A. A proteomic analysis of human bile. Mol. Cell. Proteom. 2004, 3, 715–728. [Google Scholar] [CrossRef] [Green Version]
- Farina, A.; Dumonceau, J.M.; Delhaye, M.; Frossard, J.L.; Hadengue, A.; Hochstrasser, D.F.; Lescuyer, P. A step further in the analysis of human bile proteome. J. Proteome Res. 2011, 10, 2047–2063. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Brentnall, T.A.; Chen, R. Proteomics analysis of bodily fluids in pancreatic cancer. Proteomics 2015, 15, 2705–2715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Chen, L.; Chang, H.T. Potential diagnostic and prognostic biomarkers for cholangiocarcinoma in serum and bile. Biomark. Med. 2016, 10, 613–619. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Gelrud, A.; Agarwal, B. Biliary strictures: Diagnostic considerations and approach. Gastroenterol. Rep. 2015, 3, 22–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen Canh, H.; Harada, K. Adult bile duct strictures: Differentiating benign biliary stenosis from cholangiocarcinoma. Med. Mol. Morphol. 2016, 49, 189–202. [Google Scholar] [CrossRef]
- Shanbhogue, A.K.P.; Tirumani, S.H.; Prasad, S.R.; Fasih, N.; McInnes, M. Benign biliary strictures: A current comprehensive clinical and imaging review. Am. J. Roentgenol. 2011, 197, W295–W306. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, A.A.; Krige, J.E.J.; Bornman, P.C. Biliary tract obstruction in chronic pancreatitis. HPB 2007, 9, 421–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, S.P.; Goodchild, G.; Webster, G.J.M. The endoscopist and malignant and non-malignant biliary obstruction. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 1478–1483. [Google Scholar] [CrossRef]
- Rizvi, S.; Khan, S.A.; Hallemeier, C.L.; Kelley, R.K.; Gores, G.J. Cholangiocarcinoma—Evolving concepts and therapeutic strategies. Nat. Rev. Clin. Oncol. 2017, 15, 95–111. [Google Scholar] [CrossRef] [Green Version]
- McGuigan, A.; Kelly, P.; Turkington, R.C.; Jones, C.; Coleman, H.G.; McCain, R.S. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 2018, 24, 4846–4861. [Google Scholar] [CrossRef]
- Macias, R.I.R.; Kornek, M.; Rodrigues, P.M.; Paiva, N.A.; Castro, R.E.; Urban, S.; Pereira, S.P.; Cadamuro, M.; Rupp, C.; Loosen, S.H.; et al. Diagnostic and prognostic biomarkers in cholangiocarcinoma. Liver Int. 2019, 39, 108–122. [Google Scholar] [CrossRef] [Green Version]
- Singhi, A.D.; Nikiforova, M.N.; Chennat, J.; Papachristou, G.I.; Khalid, A.; Rabinovitz, M.; Das, R.; Sarkaria, S.; Ayasso, M.S.; Wald, A.I.; et al. Integrating next-generation sequencing to endoscopic retrograde cholangiopancreatography (ERCP)-obtained biliary specimens improves the detection and management of patients with malignant bile duct strictures. Gut 2020, 69, 52–61. [Google Scholar] [CrossRef] [Green Version]
- Voigtländer, T.; Lankisch, T.O. Endoscopic diagnosis of cholangiocarcinoma: From endoscopic retrograde cholangiography to bile proteomics. Best Pract. Res. Clin. Gastroenterol. 2015, 29, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Cox, I.J.; Thillainayagam, A.V.; Bansi, D.S.; Thomas, H.C.; Taylor-Robinson, S.D. Proton and phosphorus-31 nuclear magnetic resonance spectroscopy of human bile in hepatopancreaticobiliary cancer. Eur. J. Gastroenterol. Hepatol. 2005, 17, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Ijare, O.B.; Bezabeh, T.; Albiin, N.; Arnelo, U.; Bergquist, A.; Lindberg, B.; Smith, I.C.P. Absence of glycochenodeoxycholic acid (GCDCA) in human bile is an indication of cholestasis: A 1H MRS study. NMR Biomed. 2009, 22, 471–479. [Google Scholar] [CrossRef]
- Navaneethan, U.; Gutierrez, N.G.; Venkatesh, P.G.K.; Jegadeesan, R.; Zhang, R.; Jang, S.; Sanaka, M.R.; Vargo, J.J.; Parsi, M.A.; Feldstein, A.E.; et al. Lipidomic profiling of bile in distinguishing benign from malignant biliary strictures: A single-blinded pilot study. Am. J. Gastroenterol. 2014, 109, 895–902. [Google Scholar] [CrossRef] [PubMed]
- Farina, A.; Dumonceau, J.M.; Lescuyer, P. Proteomic analysis of human bile and potential applications for cancer diagnosis. Expert Rev. Proteom. 2009, 6, 285–301. [Google Scholar] [CrossRef]
- Barbhuiya, M.A.; Sahasrabuddhe, N.A.; Pinto, S.M.; Muthusamy, B.; Singh, T.D.; Nanjappa, V.; Keerthikumar, S.; Delanghe, B.; Harsha, H.C.; Chaerkady, R.; et al. Comprehensive proteomic analysis of human bile. Proteomics 2011, 11, 4443–4453. [Google Scholar] [CrossRef]
- Lankisch, T.O.; Metzger, J.; Negm, A.A.; Vokuhl, K.; Schiffer, E.; Siwy, J.; Weismüller, T.J.; Schneider, A.S.; Thedieck, K.; Baumeister, R.; et al. Bile proteomic profiles differentiate cholangiocarcinoma from primary sclerosing cholangitis and choledocholithiasis. Hepatology 2011, 53, 875–884. [Google Scholar] [CrossRef]
- Shen, J.; Wang, W.; Wu, J.; Feng, B.; Chen, W.; Wang, M.; Tang, J.; Wang, F.; Cheng, F.; Pu, L.; et al. Comparative Proteomic Profiling of Human Bile Reveals SSP411 as a Novel Biomarker of Cholangiocarcinoma. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Lukic, N.; Visentin, R.; Delhaye, M.; Frossard, J.L.; Lescuyer, P.; Dumonceau, J.M.; Farina, A. An integrated approach for comparative proteomic analysis of human bile reveals overexpressed cancer-associated proteins in malignant biliary stenosis. Biochim. Biophys. Acta Proteins Proteom. 2014, 1844, 1026–1033. [Google Scholar] [CrossRef] [PubMed]
- Navaneethan, U.; Lourdusamy, V.; Venkatesh, P.G.K.; Willard, B.; Sanaka, M.R.; Parsi, M.A. Bile proteomics for differentiation of malignant from benign biliary strictures: A pilot study. Gastroenterol. Rep. 2015, 3, 136–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, H.; Luo, M.; Chen, J.; Zhou, Y.; Li, X.; Zhan, Y.; Shen, D.; Chen, B. Identification of TPD52 and DNAJB1 as two novel bile biomarkers for cholangiocarcinoma by iTRAQ-based quantitative proteomics analysis. Oncol. Rep. 2019, 42, 2622–2634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voigtländer, T.; Metzger, J.; Husi, H.; Kirstein, M.M.; Pejchinovski, M.; Latosinska, A.; Frantzi, M.; Mullen, W.; Book, T.; Mischak, H.; et al. Bile and urine peptide marker profiles: Access keys to molecular pathways and biological processes in cholangiocarcinoma. J. Biomed. Sci. 2020, 27, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayo, R.; Crespo, J.; Martínez-Arranz, I.; Banales, J.M.; Arias, M.; Mincholé, I.; Aller de la Fuente, R.; Jimenez-Agüero, R.; Alonso, C.; de Luis, D.A.; et al. Metabolomic-based noninvasive serum test to diagnose nonalcoholic steatohepatitis: Results from discovery and validation cohorts. Hepatol. Commun. 2018, 2, 807–820. [Google Scholar] [CrossRef] [PubMed]
- Banales, J.M.; Iñarrairaegui, M.; Arbelaiz, A.; Milkiewicz, P.; Muntané, J.; Muñoz-Bellvis, L.; La Casta, A.; Gonzalez, L.M.; Arretxe, E.; Alonso, C.; et al. Serum Metabolites as Diagnostic Biomarkers for Cholangiocarcinoma, Hepatocellular Carcinoma, and Primary Sclerosing Cholangitis. Hepatology 2019, 70, 547–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho, D.M.; Collins, K.M.; Powers, R.K.; Costello, J.C.; Collins, J.J. Next-Generation Machine Learning for Biological Networks. Cell 2018, 173, 1581–1592. [Google Scholar] [CrossRef] [Green Version]
- Perakakis, N.; Polyzos, S.A.; Yazdani, A.; Sala-Vila, A.; Kountouras, J.; Anastasilakis, A.D.; Mantzoros, C.S. Non-invasive diagnosis of non-alcoholic steatohepatitis and fibrosis with the use of omics and supervised learning: A proof of concept study. Metabolism 2019, 101, 154005. [Google Scholar] [CrossRef]
- Gerl, M.J.; Klose, C.; Surma, M.A.; Fernandez, C.; Melander, O.; Männistö, S.; Borodulin, K.; Havulinna, A.S.; Salomaa, V.; Ikonen, E.; et al. Machine learning of human plasma lipidomes for obesity estimation in a large population cohort. PLoS Biol. 2019, 17, e3000443. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, J.; Bar-Sinai, Y.; Lee, L.M.; Andrejevic, J.; Mishra, S.; Rubinstein, S.M.; Rycroft, C.H. Machine learning in a data-limited regime: Augmenting experiments with synthetic data uncovers order in crumpled sheets. Sci. Adv. 2019, 5, eaau6792. [Google Scholar] [CrossRef] [Green Version]
- Poss, A.M.; Maschek, J.A.; Cox, J.E.; Hauner, B.J.; Hopkins, P.N.; Hunt, S.C.; Holland, W.L.; Summers, S.A.; Playdon, M.C. Machine learning reveals serum sphingolipids as cholesterol-independent biomarkers of coronary artery disease. J. Clin. Investig. 2020, 130, 1363–1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deo, R.C. Machine learning in medicine. Circulation 2015, 132, 1920–1930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, C.K.; Smith, A.M.; Walsh, J.R.; Simon, A.J.; Edgar, C.; Jack, C.R.; Holtzman, D.; Russell, D.; Hill, D.; Grosset, D.; et al. Machine learning for comprehensive forecasting of Alzheimer’s Disease progression. Sci. Rep. 2019, 9, 13622. [Google Scholar] [CrossRef] [PubMed]
- Avila, M.A. Metabolomic and Proteomic Analyses of Human Bile; unpublished observations; CIMA, University of Navarra: Pamplona, Spain, 2020. [Google Scholar]
- Nagana Gowda, G.A.; Shanaiah, N.; Cooper, A.; Maluccio, M.; Raftery, D. Visualization of bile homeostasis using 1H-NMR spectroscopy as a route for assessing liver cancer. Lipids 2009, 44, 27–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weykamp, C. HbA1c: A review of analytical and clinical aspects. Ann. Lab. Med. 2013, 33, 393–400. [Google Scholar] [CrossRef] [Green Version]
- Bzdok, D.; Altman, N.; Krzywinski, M. Points of Significance: Statistics versus machine learning. Nat. Methods 2018, 15, 233–234. [Google Scholar] [CrossRef]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef] [Green Version]
- Gelman, A.; Yu-Sung, S. Arm: Data Analysis Using Regression and Multilevel/Hierarchical Models. Available online: https://cran.r-project.org/package=arm (accessed on 2 February 2020).
- Kuhn, M.; Quinlan, R. C50: C5.0 Decision Trees and Rule-Based Models. Available online: https://cran.r-project.org/ (accessed on 2 February 2020).
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S, 4th ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Chicco, D.; Rovelli, C. Computational prediction of diagnosis and feature selection on mesothelioma patient health records. PLoS ONE 2019, 14, e0208737. [Google Scholar] [CrossRef]
- Rahnemai-Azar, A.A.; Weisbrod, A.; Dillhoff, M.; Schmidt, C.; Pawlik, T.M. Intrahepatic cholangiocarcinoma: Molecular markers for diagnosis and prognosis. Surg. Oncol. 2017, 26, 125–137. [Google Scholar] [CrossRef]
- Koopmann, J.; Thuluvath, P.J.; Zahurak, M.L.; Kristiansen, T.Z.; Pandey, A.; Schulick, R.; Argani, P.; Hidalgo, M.; Iacobelli, S.; Goggins, M.; et al. Mac-2-binding protein is a diagnostic marker for biliary tract carcinoma. Cancer 2004, 101, 1609–1615. [Google Scholar] [CrossRef]
- Son, K.H.; Ahn, C.B.; Kim, H.J.; Kim, J.S. Quantitative proteomic analysis of bile in extrahepatic cholangiocarcinoma patients. J. Cancer 2020, 11, 4073–4080. [Google Scholar] [CrossRef] [PubMed]
- Farina, A.; Dumonceau, J.M.; Frossard, J.L.; Hadengue, A.; Hochstrasser, D.F.; Lescuyer, P. Proteomic analysis of human bile from malignant biliary stenosis induced by pancreatic cancer. J. Proteome Res. 2009, 8, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Zabron, A.A.; Horneffer-Van Der Sluis, V.M.; Wadsworth, C.A.; Laird, F.; Gierula, M.; Thillainayagam, A.V.; Vlavianos, P.; Westaby, D.; Taylor-Robinson, S.D.; Edwards, R.J.; et al. Elevated levels of neutrophil gelatinase-associated lipocalin in bile from patients with malignant pancreatobiliary disease. Am. J. Gastroenterol. 2011, 106, 1711–1717. [Google Scholar] [CrossRef]
- Kawase, H.; Fujii, K.; Miyamoto, M.; Kubota, K.C.; Hirano, S.; Kondo, S.; Inagaki, F. Differential LC-MS-based proteomics of surgical human cholangiocarcinoma tissues. J. Proteome Res. 2009, 8, 4092–4103. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Luo, Y.; Zhang, D.; Wang, X.; Zhang, P.; Li, H.; Ejaz, S.; Liang, S. PGK1-mediated cancer progression and drug resistance. Am. J. Cancer Res. 2019, 9, 2280–2302. [Google Scholar] [PubMed]
- Amiri, M.; Naim, H.Y. Posttranslational Processing and Function of Mucosal Maltases. J. Pediatr. Gastroenterol. Nutr. 2018, 66, S18–S23. [Google Scholar] [CrossRef]
- Lee, J.; Lee, J.; Yun, J.H.; Jeong, D.G.; Kim, J.H. DUSP28 links regulation of Mucin 5B and Mucin 16 to migration and survival of AsPC-1 human pancreatic cancer cells. Tumor Biol. 2016, 37, 12193–12202. [Google Scholar] [CrossRef]
- Mohelnikova-Duchonova, B.; Brynychova, V.; Oliverius, M.; Honsova, E.; Kala, Z.; Muckova, K.; Soucek, P. Differences in transcript levels of ABC transporters between pancreatic adenocarcinoma and nonneoplastic tissues. Pancreas 2013, 42, 707–716. [Google Scholar] [CrossRef]
- Chai, P.; Yu, J.; Ge, S.; Jia, R.; Fan, X. Genetic alteration, RNA expression, and DNA methylation profiling of coronavirus disease 2019 (COVID-19) receptor ACE2 in malignancies: A pan-cancer analysis. J. Hematol. Oncol. 2020, 13, 43. [Google Scholar] [CrossRef]
- Duan, R.D.; Hindorf, U.; Cheng, Y.; Bergenzaun, P.; Hall, M.; Hertervig, E.; Nilsson, Å. Changes of activity and isoforms of alkaline sphingomyelinase (nucleotide pyrophosphatase phosphodiesterase 7) in bile from patients undergoing endoscopic retrograde cholangiopancreatography. BMC Gastroenterol. 2014, 14, 138. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Liu, J.; Liang, X.; Chen, J.; Hong, J.; Li, L.; He, Q.; Cai, X. History and progression of Fat cadherins in health and disease. OncoTargets Ther. 2016, 9, 7337–7343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hay, D.W.; Cahalane, M.J.; Timofeyeva, N.; Carey, M.C. Molecular species of lecithins in human gallbladder bile. J. Lipid Res. 1993, 34, 759–768. [Google Scholar] [PubMed]
- Eckhardt, E.R.M.; Moschetta, A.; Renooij, W.; Goerdayal, S.S.; Van Berge-Henegouwen, G.P.; Van Erpecum, K.J. Asymmetric distribution of phosphatidylcholine and sphingomyelin between micellar and vesicular phases’: Potential implications for canalicular bile formation. J. Lipid Res. 1999, 40, 2022–2033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bikman, B.T.; Summers, S.A. Ceramides as modulators of cellular and whole-body metabolism. J. Clin. Investig. 2011, 121, 4222–4230. [Google Scholar] [CrossRef] [Green Version]
- Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 2017, 18, 33–50. [Google Scholar] [CrossRef]
- Nyberg, L.; Duan, R.D.; Axelson, J.; Nilsson, Å. Identification of an alkaline sphingomyelinase activity in human bile. Biochim. Biophys. Acta Lipids Lipid Metab. 1996, 1300, 42–48. [Google Scholar] [CrossRef]
- Duan, R.D. Alkaline sphingomyelinase (NPP7) in hepatobiliary diseases: A field that needs to be closely studied. World J. Hepatol. 2018, 10, 246–253. [Google Scholar] [CrossRef]
- Manni, M.M.; Sot, J.; Arretxe, E.; Gil-Redondo, R.; Falcón-Pérez, J.M.; Balgoma, D.; Alonso, C.; Goñi, F.M.; Alonso, A. The fatty acids of sphingomyelins and ceramides in mammalian tissues and cultured cells: Biophysical and physiological implications. Chem. Phys. Lipids 2018, 217, 29–34. [Google Scholar] [CrossRef]
- Hartmann, D.; Wegner, M.S.; Wanger, R.A.; Ferreirós, N.; Schreiber, Y.; Lucks, J.; Schiffmann, S.; Geisslinger, G.; Grösch, S. The equilibrium between long and very long chain ceramides is important for the fate of the cell and can be influenced by co-expression of CerS. Int. J. Biochem. Cell Biol. 2013, 45, 1195–1203. [Google Scholar] [CrossRef]
- Bezabeh, T.; Ijare, O.B.; Albiin, N.; Arnelo, U.; Lindberg, B.; Smith, I.C.P. Detection and quantification of d-glucuronic acid in human bile using 1H NMR spectroscopy: Relevance to the diagnosis of pancreatic cancer. Magn. Reson. Mater. Phys. Biol. Med. 2009, 22, 267–275. [Google Scholar] [CrossRef]
- Hashim AbdAlla, M.S.; Taylor-Robinson, S.D.; Sharif, A.W.; Williams, H.R.T.; Crossey, M.M.E.; Badra, G.A.; Thillainayagam, A.V.; Bansi, D.S.; Thomas, H.C.; Waked, I.A.; et al. Differences in phosphatidylcholine and bile acids in bile from Egyptian and UK patients with and without cholangiocarcinoma. HPB 2011, 13, 385–390. [Google Scholar] [CrossRef] [Green Version]
- Sun, K.; Chen, S.; Xu, J.; Li, G.; He, Y. The prognostic significance of the prognostic nutritional index in cancer: A systematic review and meta-analysis. J. Cancer Res. Clin. Oncol. 2014, 140, 1537–1549. [Google Scholar] [CrossRef] [PubMed]
- Van Helvoort, A.; Smith, A.J.; Sprong, H.; Fritzsche, I.; Schinkel, A.H.; Borst, P.; Van Meer, G. MDR1 P-glycoprotein is a lipid translocase of broad specificity, while MDR3 P-glycoprotein specifically translocates phosphatidylcholine. Cell 1996, 87, 507–517. [Google Scholar] [CrossRef] [Green Version]
- Mutanen, A.; Lohi, J.; Heikkilä, P.; Jalanko, H.; Pakarinen, M.P. Liver Inflammation Relates to Decreased Canalicular Bile Transporter Expression in Pediatric Onset Intestinal Failure. Ann. Surg. 2018, 268, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Ehling, J.; Tacke, F. Role of chemokine pathways in hepatobiliary cancer. Cancer Lett. 2016, 379, 173–183. [Google Scholar] [CrossRef]
- Geier, A.; Wagner, M.; Dietrich, C.G.; Trauner, M. Principles of hepatic organic anion transporter regulation during cholestasis, inflammation and liver regeneration. Biochim. Biophys. Acta Mol. Cell Res. 2007, 1773, 283–308. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Ishigami, M.; Nagao, K.; Hanada, K.; Kono, N.; Arai, H.; Matsuo, M.; Kioka, N.; Ueda, K. ABCB4 exports phosphatidylcholine in a sphingomyel-independent manner. J. Lipid Res. 2015, 56, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Sonkar, K.; Ayyappan, V.; Tressler, C.M.; Adelaja, O.; Cai, R.; Cheng, M.; Glunde, K. Focus on the glycerophosphocholine pathway in choline phospholipid metabolism of cancer. NMR Biomed. 2019, 32. [Google Scholar] [CrossRef]
- Braverman, N.E.; Moser, A.B. Functions of plasmalogen lipids in health and disease. Biochim. Biophys. Acta Mol. Basis Dis. 2012, 1822, 1442–1452. [Google Scholar] [CrossRef] [Green Version]
- Bose, S.; Ramesh, V.; Locasale, J.W. Acetate Metabolism in Physiology, Cancer, and Beyond. Trends Cell Biol. 2019, 29, 695–703. [Google Scholar] [CrossRef]
- Madhu, B.; Narita, M.; Jauhiainen, A.; Menon, S.; Stubbs, M.; Tavaré, S.; Narita, M.; Griffiths, J.R. Metabolomic changes during cellular transformation monitored by metabolite–metabolite correlation analysis and correlated with gene expression. Metabolomics 2015, 11, 1848–1863. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y. Bin Creatine kinase in cell cycle regulation and cancer. Amino Acids 2016, 48, 1775–1784. [Google Scholar] [CrossRef] [PubMed]
- Pietzke, M.; Meiser, J.; Vazquez, A. Formate metabolism in health and disease. Mol. Metab. 2020, 33, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Boroughs, L.K.; Deberardinis, R.J. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 2015, 17, 351–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [Green Version]
- Permert, J.; Ihse, I.; Jorfeldt, L.; von Schennck, H.; Arnqvist, H.; Larsson, J. Pancreatic cancer is associated with impaired glucose metabolism. Eur. J. Surg. 1993, 159, 101–107. [Google Scholar]
- Roeyen, G.; Jansen, M.; Chapelle, T.; Bracke, B.; Hartman, V.; Ysebaert, D.; De Block, C. Diabetes mellitus and pre-diabetes are frequently undiagnosed and underreported in patients referred for pancreatic surgery. A prospective observational study. Pancreatology 2016, 16, 671–676. [Google Scholar] [CrossRef]
- Mancinelli, R.; Olivero, F.; Carpino, G.; Overi, D.; Rosa, L.; Lepanto, M.S.; Cutone, A.; Franchitto, A.; Alpini, G.; Onori, P.; et al. Role of lactoferrin and its receptors on biliary epithelium. BioMetals 2018, 31, 369–379. [Google Scholar] [CrossRef]
- Thomas, D.G.; Robinson, D.N. The fifth sense: Mechanosensory regulation of alpha-actinin-4 and its relevance for cancer metastasis. Semin. Cell Dev. Biol. 2017, 71, 68–74. [Google Scholar] [CrossRef]
- Zang, Z.J.; Cutcutache, I.; Poon, S.L.; Zhang, S.L.; Mcpherson, J.R.; Tao, J.; Rajasegaran, V.; Heng, H.L.; Deng, N.; Gan, A.; et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat. Genet. 2012, 44, 570–574. [Google Scholar] [CrossRef]
- Martínez-Arranz, I.; Mayo, R.; Pérez-Cormenzana, M.; Mincholé, I.; Salazar, L.; Alonso, C.; Mato, J.M. Enhancing metabolomics research through data mining. J. Proteom. 2015, 127, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Monte, M.J.; Martinez-Diez, M.C.; El-Mir, M.Y.; Mendoza, M.E.; Bravo, P.; Bachs, O.; Marin, J.J.G. Changes in the pool of bile acids in hepatocyte nuclei during rat liver regeneration. J. Hepatol. 2002, 36, 534–542. [Google Scholar] [CrossRef]
- Nytofte, N.S.; Serrano, M.A.; Monte, M.J.; Gonzalez-Sanchez, E.; Tumer, Z.; Ladefoged, K.; Briz, O.; Marin, J.J.G. A homozygous nonsense mutation (c.214C→A) in the biliverdin reductase alpha gene (BLVRA) results in accumulation of biliverdin during episodes of cholestasis. J. Med. Genet. 2011, 48, 219–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholson, J.K.; Foxall, P.J.D.; Spraul, M.; Farrant, R.D.; Lindon, J.C. 750 MHz 1H and 1H-13C NMR Spectroscopy of Human Blood Plasma. Anal. Chem. 1995, 67, 793–811. [Google Scholar] [CrossRef]
- Jacob, D.; Deborde, C.; Lefebvre, M.; Maucourt, M.; Moing, A. NMRProcFlow: A graphical and interactive tool dedicated to 1D spectra processing for NMR-based metabolomics. Metabolomics 2017, 13, 36. [Google Scholar] [CrossRef] [Green Version]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in1H NMR metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef] [PubMed]
- Markley, J.L.; Ulrich, E.L.; Berman, H.M.; Henrick, K.; Nakamura, H.; Akutsu, H. BioMagResBank (BMRB) as a partner in the Worldwide Protein Data Bank (wwPDB): New policies affecting biomolecular NMR depositions. J. Biomol. NMR 2008, 40, 153–155. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Ramos-Fernández, A.; Paradela, A.; Navajas, R.; Albar, J.P. Generalized method for probability-based peptide and protein identification from tandem mass spectrometry data and sequence database searching. Mol. Cell. Proteom. 2008, 7, 1748–1754. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Serra, P.; Marcilla, M.; Villanueva, A.; Ramos-Fernandez, A.; Palau, A.; Leal, L.; Wahi, J.E.; Setien-Baranda, F.; Szczesna, K.; Moutinho, C.; et al. A DERL3-associated defect in the degradation of SLC2A1 mediates the Warburg effect. Nat. Commun. 2014, 5, 3608. [Google Scholar] [CrossRef] [Green Version]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Linares, M.; Hewapathinara, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucl. Acid Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Curran-Everett, D. Explorations in statistics: Permutation methods. Am. J. Physiol. Adv. Physiol. Educ. 2012, 36, 181–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arima, K.; Lau, M.C.; Zhao, M.; Haruki, K.; Kosumi, K.; Mima, K.; Gu, M.; Väyrynen, J.P.; Twombly, T.S.; Baba, Y.; et al. Metabolic Profiling of Formalin-Fixed Paraffi n-Embedded Tissues Discriminates Normal Colon from Colorectal Cancer. Mol. Cancer Res. 2020, 18, 883–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, R.A. The use of multiple measurements in taxonomic problems. Ann. Eugen. 1936, 7, 179–188. [Google Scholar] [CrossRef]
- R Core Team. The R Project for Statistical Computing. Available online: http://www.r-project.org (accessed on 2 February 2020).
- Kowaric, A.; Templ, M. Imputation with the {R} Package {VIM}. J. Stat. Softw. 2016, 74, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z. Missing data exploration: Highlighting graphical presentation of missing pattern. Ann. Transl. Med. 2015, 3, 356. [Google Scholar] [CrossRef]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Andronesi, O.C.; Blekas, K.D.; Mintzopoulos, D.; Astrakas, L.; Black, P.M.; Tzika, A.A. Molecular classification of brain tumor biopsies using solid-state magic angle spinning proton magnetic resonance spectroscopy and robust classifiers. Int. J. Oncol. 2008, 33, 1017–1025. [Google Scholar] [CrossRef]
- Weng, S.F.; Reps, J.; Kai, J.; Garibaldi, J.M.; Qureshi, N. Can machine-learning improve cardiovascular risk prediction using routine clinical data? PLoS ONE 2017, 12, e0174944. [Google Scholar] [CrossRef] [Green Version]
- Beck, M.W. NeuralNetTools: Visualization and Analysis Tools for Neural Networks. J. Stat. Softw. 2018, 85, 1. [Google Scholar] [CrossRef]
- Semba, R.D.; Zhang, P.; Adelnia, F.; Sun, K.; Gonzalez-Freire, M.; Salem, N.; Brennan, N.; Spencer, R.G.; Fishbein, K.; Khadeer, M.; et al. Low plasma lysophosphatidylcholines are associated with impaired mitochondrial oxidative capacity in adults in the Baltimore Longitudinal Study of Aging. Aging Cell 2019, 18, e12915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.J.; Cho, K.J.; Oh, S. Development of machine learning models for diagnosis of glaucoma. PLoS ONE 2017, 12, e0177726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.H.; Han, K. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology 2018, 286, 800–809. [Google Scholar] [CrossRef] [PubMed]
| Variables | Benign Biliary Conditions (n = 36) | CCA (n = 36) | PDAC (n = 57) | p Value * |
|---|---|---|---|---|
| Age, median (years) ± SD | 66 ± 19 | 74 ± 12 | 71 ± 12 | ap = 0.05, b p = 0.09 |
| Gender (Male/Female) | 19/17 | 17/19 | 25/32 | p = 0.718 |
| Location of biliary stenosis (Distal/Hilar/Intrahepatic) | 10/0/1 | 18/15/3 | 57/0/0 | |
| Operated stenosis ** | 1 (9.1%) | 14 (38.9%) | 16 (28%) | |
| Stage IV (AJCC Pronostic Group ***) | NA | 8 (22.2%) | 15 (26.3%) | |
| Body Mass Index (kg/m2) | 27.28 ± 4.56 | 25.26 ± 4.65 | 25.86 ± 4.96 | ap = 0.067, b p = 0.169 |
| Bilirrubin (mg/dL) | 3.18 ± 3.10 | 9.05 ± 7.78 | 10.79 ± 7.11 | ap = 0.00019, b p = 0.00000037 |
| Albumin (g/dL) | 3.69 ± 0.47 | 3.29 ± 0.57 | 3.46 ± 0.47 | ap = 0.0029, b p = 0.029 |
| GGT (U/L) | 609 ± 517 | 1013 ± 678 | 1116 ± 724 | ap = 0.0078, b p = 0.00083 |
| INR | 1.13 ± 0.17 | 1.14 ± 0.22 | 1.13 ± 0.15 | ap = 0.8, b p = 0.98 |
| Total cholesterol (mg/dL) | 171 ± 48 | 225 ± 82 | 233 ± 107 | ap = 0.0018, b p = 0.0026 |
| Triglycerides (mg/dL) | 138 ± 81 | 169 ± 105 | 178 ± 81 | ap = 0.187, b p = 0.031 |
| PNI **** | 44.80 ± 6.74 | 41.41 ± 6.81 | 41.82 ± 5.95 | ap = 0.042, b p = 0.033 |
| High CA19-9 (>37 U/L) ***** | 10 (27.8%) | 24 (66.7%) | 46 (80.7%) | ap = 0.578, b p = 0.065 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urman, J.M.; Herranz, J.M.; Uriarte, I.; Rullán, M.; Oyón, D.; González, B.; Fernandez-Urién, I.; Carrascosa, J.; Bolado, F.; Zabalza, L.; et al. Pilot Multi-Omic Analysis of Human Bile from Benign and Malignant Biliary Strictures: A Machine-Learning Approach. Cancers 2020, 12, 1644. https://doi.org/10.3390/cancers12061644
Urman JM, Herranz JM, Uriarte I, Rullán M, Oyón D, González B, Fernandez-Urién I, Carrascosa J, Bolado F, Zabalza L, et al. Pilot Multi-Omic Analysis of Human Bile from Benign and Malignant Biliary Strictures: A Machine-Learning Approach. Cancers. 2020; 12(6):1644. https://doi.org/10.3390/cancers12061644
Chicago/Turabian StyleUrman, Jesús M., José M. Herranz, Iker Uriarte, María Rullán, Daniel Oyón, Belén González, Ignacio Fernandez-Urién, Juan Carrascosa, Federico Bolado, Lucía Zabalza, and et al. 2020. "Pilot Multi-Omic Analysis of Human Bile from Benign and Malignant Biliary Strictures: A Machine-Learning Approach" Cancers 12, no. 6: 1644. https://doi.org/10.3390/cancers12061644







