Circulating miRNAs in Small Extracellular Vesicles Secreted by a Human Melanoma Xenograft in Mouse Brains
Abstract
:1. Introduction
2. Results
3. Discussion
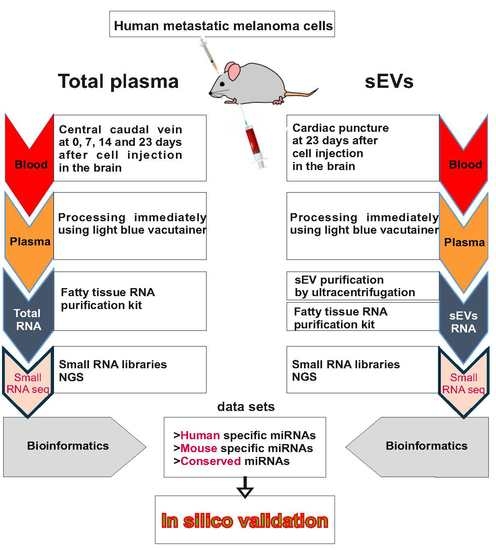
4. Materials and Methods
4.1. Cell Line Maintenance
4.2. In Vivo Intracranial Xenografts
4.3. Ethical Review Procedure
4.4. Histochemistry
4.5. sEV Isolation and Purification from Cell Cultures
4.6. sEV Isolation and Purification from Blood Plasma
4.7. Dynamic Light Scattering (DLS) and Zeta Potential
4.8. Light and Electron Microscopy
4.9. Flow Cytometry
4.10. Total RNA Preparation
4.11. Plasma and sEV-Carried Total Small RNA Sequencing
4.12. miRNA Bioinformatics Analysis of Mouse Plasma-Derived Samples
4.13. Identification of miRNA Target Genes and Functional Enrichment Analysis
4.14. Analysis of EV-Associated miRNAs in Human Metastatic Melanoma Cases
4.15. Analysis of miRNAs Expressed in Xenograft Tissue of M14 Metastatic Melanoma Cells
4.16. Data Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Disibio, G.; French, S.W. Metastatic patterns of cancers: Results from a large autopsy study. Arch. Pathol. Lab. Med. 2008, 132, 931–939. [Google Scholar] [PubMed]
- Barnholtz-Sloan, J.S.; Sloan, A.E.; Davis, F.G.; Vigneau, F.D.; Lai, P. Sawaya RE. Incidence proportions of brain metastases in patients diagnosed (1973 to 2001) in the Metropolitan Detroit Cancer Surveillance System. J. Clin. Oncol. 2004, 22, 2865–2872. [Google Scholar] [CrossRef]
- Schouten, L.J.; Rutten, J.; Huveneers, H.A.; Twijnstra, A. Incidence of brain metastases in a cohort of patients with carcinoma of the breast, colon, kidney, and lung and melanoma. Cancer 2002, 94, 2698–2705. [Google Scholar] [CrossRef] [PubMed]
- Gibney, G.T.; Forsyth, P.A.; Sondak, V.K. Melanoma in the brain: Biology and therapeutic options. Melanoma Res. 2012, 22, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Venur, V.A.; Funchain, P.; Kotecha, R.; Chao, S.T.; Ahluwalia, M.S. Changing Treatment Paradigms for Brain Metastases from Melanoma-Part 1: Diagnosis, Prognosis, Symptom Control, and Local Treatment. Oncology 2017, 31, 602–606. [Google Scholar]
- Zakrzewski, J.; Geraghty, L.N.; Rose, A.E.; Christos, P.J.; Mazumdar, M.; Polsky, D.; Shapiro, R.; Berman, R.; Darvishian, F.; Hernando, E.; et al. Clinical Variables and Primary Tumor Characteristics Predictive of the Development of Melanoma Brain Metastases and Post-Brain Metastases Survival. Cancer 2011, 117, 1711–1720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedikian, A.Y.; Wei, C.; Detry, M.; Kim, K.B.; Papadopoulos, N.E.; Hwu, W.J.; Homsi, J.; Davies, M.; McIntyre, S.; Hwu, P. Predictive Factors for the Development of Brain Metastasis in Advanced Unresectable Metastatic Melanoma. Am. J. Clin. Oncol. 2010, 34, 603–610. [Google Scholar] [CrossRef]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef]
- Gurtan, A.; Sharp, P.A. The role of miRNAs in regulating gene expression networks. J. Mol. Biol. 2013, 425, 3582–3600. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [Green Version]
- Valenti, M.T.; Dalle Carbonare, L.; Mottes, M. Role of microRNAs in progenitor cell commitment and osteogenic differentiation in health and disease. Int. J. Mol. Med. 2018, 41, 2441–2449. [Google Scholar] [CrossRef] [Green Version]
- Bennett, P.E.; Bemis, L.; Norris, D.A.; Shellman, Y.G. MiR in melanoma development: miRNAs and acquired hallmarks of cancer in melanoma. Physiol. Genom. 2013, 45, 1049–1059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thyagarajana, A.; Tsaib, K.Y.; Sahua, R.P. MicroRNA heterogeneity in melanoma progression. Semin. Cancer Biol. 2019, 59, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Gajos-Michniewicz, A.; Czyz, M. Role of miRNAs in Melanoma Metastasis. Cancers 2019, 11, 326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thuma, F.; Zoller, M. Outsmart tumor exosomes to steal the cancer initiating cell its niche. Semin. Cancer Biol. 2014, 28, 39–50. [Google Scholar] [CrossRef]
- Taverna, S.; Amodeo, V.; Saieva, L.; Russo, A.; Giallombardo, M.; De, L.G.; Alessandro, R. Exosomal shuttling of miR-126 in endothelial cells modulates adhesive and migratory abilities of chronic myelogenous leukemia cells. Mol. Cancer 2014, 13, 169. [Google Scholar] [CrossRef] [Green Version]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat. Rev. Clin Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Kahlert, C.; Kalluri, R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J. Mol. Med. 2013, 91, 431–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kharaziha, P.; Ceder, S.; Li, Q.; Panaretakis, T. Tumor cell-derived exosomes: A message in a bottle. Biochim. Biophys. Acta 2012, 1826, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Martins, T.S.; Catita, J.; Rosa, I.M.; da Cruz E Silva, O.A.B.; Henriques, A.G. Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLoS ONE 2018, 13, e0198820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witwer, K.W.; Halushka, M.K. Toward the promise of microRNAs–Enhancing reproducibility and rigor in microRNA research. RNA Biol. 2016, 13, 1103–1116. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Dindorf, J.; Eberhardt, M.; Lai, X.; Ostalecki, C.; Koliha, N.; Gross, S.; Blume, K.; Bruns, H.; Wild, S.; et al. Innate extracellular vesicles from melanoma patients suppress β-catenin in tumor cells by miRNA-34a. Life Sci. All. 2019, 2, e201800205. [Google Scholar] [CrossRef]
- Gowda, R.; Robertson, B.M.; Iyer, S.; Barry, J.; Dinavahi, S.S.; Robertson, G.P. The Role of Exosomes in Metastasis and Progression of Melanoma. Cancer Treat. Rev. 2020, 85, 101975. [Google Scholar] [CrossRef]
- Carpi, S.; Polini, B.; Fogli, S.; Podestà, A.; Ylösmäki, E.; Cerullo, V.; Romanini, A.; Nieri, P. Circulating microRNAs as biomarkers for early diagnosis of cutaneous melanoma. Exp. Rev. Mol. Diagn. 2019, 20, 19–30. [Google Scholar] [CrossRef]
- Margue, C.; Reinsbach, S.; Philippidou1, D.; Beaume, N.; Walters, C.; Schneider, J.G.; Nashan, D.; Behrmann, I.; Kreis, S. Comparison of a healthy miRNome with melanoma patient miRNomes: Are microRNAs suitable serum biomarkers for cancer? Oncotarget 2015, 6, 12110–12127. [Google Scholar] [CrossRef] [Green Version]
- Cesi, G.; Walbrecq, G.; Margue, C.; Kreis, S. Transferring intercellular signals and traits between cancer cells: Extracellular vesicles as “homing pigeons”. Cell Comm. Sign. 2016, 14, 13. [Google Scholar] [CrossRef] [Green Version]
- Milella, M.; Falcone, I.; Conciatori, F.; Matteoni, S.; Sacconi, A.; De Luca, T.; Bazzichetto, C.; Corbo, V.; Simbolo, M.; Sperduti, I.; et al. PTEN status is a crucial determinant of the functional outcome of combined MEK and mTOR inhibition in cancer. Sci. Rep. 2017, 7, 43013. [Google Scholar] [CrossRef] [Green Version]
- Kircher, D.A.; Silvis, M.R.; Cho, J.H.; Holmen, S.L. Melanoma Brain Metastasis: Mechanisms, Models, and Medicine. Int. J. Mol. Sci. 2016, 17, 1468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slattery, M.L.; Mullany, L.E.; Sakoda, L.C.; Wolff, R.K.; Stevens, J.R.; Samowitz, W.S.; Herrick, J.S. The PI3K/AKT signaling pathway: Associations of miRNAs with dysregulated gene expression in colorectal cancer. Mol. Carcinog. 2018, 57, 243–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.F.; Lee, C.Y.; Lai, L.C.; Tsai, M.H.; Lu, T.P.; Chuang, E.Y. CellExpress: A comprehensive microarray-based cancer cell line and clinical sample gene expression analysis online system. Database 2018, 2018, 1–11. [Google Scholar] [CrossRef]
- Iyengar, B.; Singh, A.V. Patterns of neural differentiation in melanomas. J. Biomed. Sci. 2010, 17, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Khordadmehr, M.; Shahbazi, R.; Sadreddini, S.; Baradaran, B. miR-193: A new weapon against cancer. J. Cell. Physiol. 2019, 234, 16861–16872. [Google Scholar] [CrossRef]
- Yang, F.; Xiong, H.; Duan, L.; Li, Q.; Li, X.; Zhou, Y. MiR-1246 Promotes Metastasis and Invasion of A549 cells by Targeting GSK-3β–Mediated Wnt/β-Catenin Pathway. Cancer Res. Treat. 2019, 51, 1420–1429. [Google Scholar] [CrossRef] [Green Version]
- Ni, J.-S.; Zheng, H.; Huang, Z.-P.; Hong, Y.-G.; Ou, Y.-L.; Tao, Y.-P.; Wang, M.-C.; Wang, Z.-G.; Yang, Y.; Zhou, W.-P. MicroRNA-197-3p acts as a prognostic marker and inhibits cell invasion in hepatocellular carcinoma. Oncol. Lett. 2019, 17, 2317–2327. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Shetti, D.; Fan, C.; Wei, K. miR-29b-3p promotes progression of MDA-MB-231 triple-negative breast cancer cells through downregulating TRAF3. Biol. Res. 2019, 52, 38. [Google Scholar] [CrossRef] [Green Version]
- Ying, X.; Zhang, W.; Fang, M.; Zhang, W.; Wang, C.; Han, L. miR-345-5p regulates proliferation, cell cycle, and apoptosis of acute myeloid leukemia cells by targeting AKT2. J. Cell. Biochem. 2018, 120, 1620–1629. [Google Scholar] [CrossRef]
- Wu, K.; Yang, L.; Chen, J.; Zhao, H.; Wang, J.; Xu, S.; Huang, Z. miR-362-5p inhibits proliferation and migration of neuroblastoma cells by targeting phosphatidylinositol 3-kinase-C2b. FEBS Lett. 2015, 589, 1911–1919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazar, J.; Qi, F.; Lee, B.; Marchica, J.; Govindarajan, S.; Shelley, J.; Li, J.-L.; Ray, A.; Perera, R.J. MicroRNA 211 Functions as a Metabolic Switch in Human Melanoma Cells. Mol. Cell. Biol. 2016, 36, 1090–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fite, K.; Gomez-Cambronero, X.J. Down-regulation of MicroRNAs (MiRs) 203, 887, 3619 and 182 Prevents Vimentin-triggered, Phospholipase D (PLD)-mediated Cancer Cell Invasion. J. Biol. Chem. 2016, 291, 719–730. [Google Scholar] [CrossRef] [Green Version]
- Ohshima, K.; Inoue, K.; Fujiwara, A.; Hatakeyama, K.; Kanto, K.; Watanabe, Y.; Muramatsu, K.; Fukuda, Y.; Ogura, S.I.; Yamaguchi, K.; et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS ONE 2010, 5, e13247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostenfeld, M.S.; Jeppesen, D.K.; Laurberg, J.R.; Boysen, A.T.; Bramsen, J.B.; Primdal-Bengtson, B.; Hendrix, A.; Lamy, P.; Dagnaes-Hansen, F.; Rasmussen, M.H.; et al. Cellular Disposal of miR23b by RAB27-Dependent Exosome Release Is Linked to Acquisition of Metastatic Properties. Cancer Res. 2014, 74, 5758–5771. [Google Scholar] [CrossRef] [Green Version]
- Igaz, I.; Igaz, P. Tumor surveillance by circulating microRNAs: A hypothesis. Cell Mol. Life Sci. 2014, 71, 4081–4087. [Google Scholar] [CrossRef] [Green Version]
- Soung, Y.H.; Nguyen, T.; Cao, H.; Lee, J.; Chung, J. Emerging roles of exosomes in cancer invasion and metastasis. BMB Rep. 2016, 49, 18–25. [Google Scholar] [CrossRef] [Green Version]
- Greening, D.W.; Gopal, S.K.; Mathias, R.A.; Liu, L.; Sheng, J.; Zhu, H.J.; Simpson, R.J. Emerging roles of exosomes during epithelial–mesenchymal transition and cancer progression. Semin. Cell Devel. Biol. 2015, 40, 60–71. [Google Scholar] [CrossRef]
- Zhang, H.G.; Grizzle, W.E. Exosomes: A novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. Am. J. Pathol. 2014, 184, 28–41. [Google Scholar] [CrossRef] [Green Version]
- Palmirotta, R.; Lovero, D.; Cafforio, P.; Felici, C.; Mannavola, F.; Pellè, E.; Quaresmini, D.; Tucci, M.; Silvestris, F. Liquid biopsy of cancer: A multimodal diagnostic tool in clinical oncology. Ther. Adv. Med. Oncol. 2018, 10, 1–24. [Google Scholar] [CrossRef]
- Cheng, L.; Sharples, R.A.; Scicluna, B.J.; Hill, A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 2014, 3, 23743. [Google Scholar] [CrossRef] [PubMed]
- Georgiadis, A.; Tschernutter, M.; Bainbridge, J.W.; Balaggan, K.S.; Mowat, F.; West, E.L.; Munro, P.M.; Thrasher, A.J.; Matter, K.; Balda, M.S.; et al. The Tight Junction Associated Signalling Proteins ZO-1 and ZONAB Regulate Retinal Pigment Epithelium Homeostasis in Mice. PLoS ONE 2010, 5, e15730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paris, L.; Tonutti, L.; Vannini, C.; Bazzoni, G. Structural organization of the tight junctions. Biochim. Biophys. Acta 2008, 1778, 646–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckler, M.D.; Higginbotham, J.N.J.; Franklin, L.; Ham, A.-J.; Halvey, P.J.; Imasuen, I.E.; Whitwell, C.; Li, M.; Liebler, D.C.; Coffey, R.J. Proteomic Analysis of Exosomes from Mutant KRAS Colon Cancer Cells Identifies Intercellular Transfer of Mutant KRAS. Mol. Cell. Proteom. 2013, 12, 343–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.E.; Tan, H.S.; Datta, A.R.; Lai, C.; Zhang, H.; Meng, W.; Lim, S.K.; Sze, S.K. Hypoxic Tumor Cell Modulates Its Microenvironment to Enhance Angiogenic and Metastatic Potential by Secretion of Proteins and Exosomes. Mol. Cell. Proteom. 2010, 9, 1085–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemcke, H.; David, R. Potential mechanisms of microRNA mobility. Traffic 2018, 19, 910–917. [Google Scholar] [CrossRef]
- Zong, L.; Zhu, Y.; Liang, R.; Zhaoa, H.-B. Gap junction mediated miRNA intercellular transfer and gene regulation: A novel mechanism for intercellular genetic communication. Sci. Rep. 2016, 6, 19884. [Google Scholar] [CrossRef] [Green Version]
- Arcega, R.; Yong, W.H.; Xu, H. Malignant melanoma mimicking giant cell variant of glioblastoma multiforme: A case report and review of literature. Int. J. Clin. Exper. Pathol. 2015, 8, 5929–5933. [Google Scholar]
- Romano, R.C.; Carter, J.M.; Folpe, A.L. Aberrant intermediate filament and synaptophysin expression is a frequent event in malignant melanoma: An immunohistochemical study of 73 cases. Mod. Pathol. 2015, 28, 1033–1042. [Google Scholar] [CrossRef] [Green Version]
- Yi, J.; Jin, L.; Chen, J.; Feng, B.; He, Z.; Chen, L.; Song, H. MiR-375 suppresses invasion and metastasis by direct targeting of SHOX2 in esophageal squamous cell carcinoma. Acta Biochim. Biophys. Sin. 2017, 49, 159–169. [Google Scholar] [CrossRef] [Green Version]
- Pascut, D.; Krmac, H.; Gilardi, F.; Patti, R.; Calligaris, R.; Crocè, L.S.; Tiribelli, C. A comparative characterization of the circulating miRNome in whole blood and serum of HCC patients. Sci. Rep. 2019, 9, 8265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, T.; Li, H.; Chen, T.; Ren, H.; Shi, P.; Chen, M. LncRNA MALAT1 Depressed Chemo-Sensitivity of NSCLC Cells through Directly Functioning on miR-197-3p/p120 Catenin Axis. Mol. Cells 2019, 42, 270–283. [Google Scholar] [PubMed]
- Qi, Y.; Zha, W.; Zhang, W. Exosomal miR-660-5p promotes tumor growth and metastasis in non-small cell lung cancer. J. Balkan Union Oncol. 2019, 24, 599–607. [Google Scholar]
- Li, Y.; Liu, Y.; Ren, J.; Deng, S.; Yi, G.; Guo, M.; Shu, S.; Zhao, L.; Peng, Y.; Qi, S. miR-1268a regulates ABCC1 expression to mediate temozolomide resistance in glioblastoma. J. Neurooncol. 2018, 138, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Ahn, J.H.; Lee, M. Upregulation of MicroRNA-1246 Is Associated with BRAF Inhibitor Resistance in Melanoma Cells with Mutant BRAF. Cancer Res. Treat. 2017, 49, 947–959. [Google Scholar] [CrossRef] [Green Version]
- Wei, F.; Cao, C.; Xu, X.; Wang, J. Diverse functions of miR-373 in cancer. J. Translat. Med. 2015, 13, 162. [Google Scholar] [CrossRef] [Green Version]
- Gargiulo, E.; Paggetti, J.; Moussay, E. Hematological Malignancy-Derived Small Extracellular Vesicles and Tumor Microenvironment: The Art of Turning Foes into Friends. Cells 2019, 8, 511. [Google Scholar] [CrossRef] [Green Version]
- Hannafon, B.N.; Ding, W.Q. Intercellular communication by exosome-derived microRNAs in cancer. Int. J. Mol. Sci. 2013, 14, 14240–14269. [Google Scholar] [CrossRef] [Green Version]
- Skog, J.; Würdinger, T.; Van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Steenbeek, S.C.; Pham, T.V.; de Ligt, J.; Zomer, A.; Knol, J.C.; Piersma, S.R.; Schelfhorst, T.; Huisjes, R.; Schiffelers, R.M.; Cuppen, E.; et al. Cancer cells copy migratory behavior and exchange signaling networks via extracellular vesicles. EMBO J. 2018, 37, e98357. [Google Scholar] [CrossRef]
- Wang, W.; Yang, J.; Yu, F.; Li, W.; Wang, L.; Zou, H.; Long, X. MicroRNA-122-3p inhibits tumor cell proliferation and induces apoptosis by targeting Forkhead box O in A549 cells. Oncol. Lett. 2018, 15, 2695–2699. [Google Scholar] [CrossRef]
- Matsumoto, A.; Takahashi, Y.; Chang, H.-Y.; Wu, Y.-W.; Yamamoto, A.; Ishihama, Y.; Takakura, Y. Blood concentrations of small extracellular vesicles are determined by a balance between abundant secretion and rapid clearance. JEV 2019, 9, 1696517. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Wang, L. Regulation of microRNA expression and function by nuclear receptor signaling. Cell Biosci. 2011, 1, 31. [Google Scholar] [CrossRef] [Green Version]
- Benes, V.; Collier, P.; Kordes, C.; Stolte, J.; Rausch, T.; Muckentaler, M.U.; Häussinger, D.; Castoldi, M. Identification of cytokine-induced modulation of microRNA expression and secretion as measured by a novel microRNA specific qPCR assay. Sci. Rep. 2015, 5, 11590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández Esmerats, J.; Heath, J.; Jo, H. Shear sensitive genes in aortic valve endothelium. Antioxid. Redox Signal. 2016, 25, 401–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riemann, A.; Reime, S.; Thews, O. Acidic extracellular environment affects miRNA expression in tumors in vitro and in vivo. Int. J. Cancer 2019, 144, 1609–1618. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinf. 2015, 13, 17–24. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.; Njauw, C.-N.; Reddy, B.Y.; Ji, Z.; Rajadurai, A.; Klebanov, N.; Tsao, H. Growth suppression by dual BRAF(V600E) and NRAS(Q61) oncogene expression is mediated by SPRY4 in melanoma. Oncogene 2019, 38, 3504–3520. [Google Scholar] [CrossRef]
- Petti, C.; Molla, A.; Vegetti, C.; Ferrone, S.; Anichini, A.; Sensi, M. Coexpression of NRASQ61R and BRAFV600E in human melanoma cells activates senescence and increases susceptibility to cell-mediated cytotoxicity. Cancer Res. 2006, 66, 6503–6511. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Zhou, J.; Mi, J.; Ma, K.; Fan, Y.; Ning, J.; Wang, C.; Wei, X.; Zhao, H.; Li, E. HOXD10 acts as a tumor-suppressive factor via inhibition of the RHOC/AKT/MAPK pathway in human cholangiocellular carcinoma. Oncol. Rep. 2015, 34, 1681–1691. [Google Scholar] [CrossRef] [Green Version]
- Lakshmanachetty, S.; Balaiya, V.; High, W.A.; Koster, M.I. Loss of TP63 promotes the metastasis of head and neck squamous cell carcinoma by activating MAPK and STAT3 signaling. Mol. Cancer Res. 2019, 17, 1279–1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexius-Lindgren, M.; Andersson, E.; Lindstedt, I.; Engström, W. The RECK gene and biological malignancy—Its significance in angiogenesis and inhibition of matrix metalloproteinases. Anticancer Res. 2014, 34, 3867–3873. [Google Scholar] [PubMed]
- Zhang, G.; Cheng, Y.; Chen, G.; Tang, Y.; Ardekani, G.; Rotte, A.; Martinka, M.; McElwee, K.; Xu, X.; Wang, Q.; et al. Loss of tumor suppressors KAI1 and p27 identifies a unique subgroup of primary melanoma patients with poor prognosis. Oncotarget 2015, 6, 23026–32305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, T.I.; Hsu, C.H.; Lee, K.H.; Lin, J.T.; Chen, C.S.; Chang, K.C.; Su, C.Y.; Hsiao, M.; Lu, P.J. MicroRNA-18a is elevated in prostate cancer and promotes tumorigenesis through suppressing STK4 in vitro and in vivo. Oncogenesis 2014, 3, e99. [Google Scholar] [CrossRef] [Green Version]
- Pusapati, R.V.; Weaks, R.L.; Rounbehler, R.J.; McArthur, M.J.; Johnson, D.G. E2F2 Suppresses Myc-induced Proliferation and Tumorigenesis. Mol. Carcinogen. 2010, 49, 152–156. [Google Scholar] [CrossRef] [Green Version]
- Johnson, D.G.; DeGregori, J. Putting the Oncogenic and Tumor Suppressive Activities of E2F into Context. Curr. Mol. Med. 2006, 6, 731–738. [Google Scholar]
- Li, J.; Liu, X.; Li, C.; Wang, W. miR-224-5p inhibits proliferation, migration, and invasion by targeting PIK3R3/AKT3 in uveal melanoma. J. Cell. Biochem. 2019, 120, 12412–12421. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, L.S.; Chen, G.; Feng, Z.B. Potential Targets and Clinical Value of MiR-224-5p in Cancers of the Digestive Tract. Cell. Physiol. Biochem. 2017, 44, 682–700. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.J.; Zhou, H.; Xiao, H.X.; Li, Y.; Zhou, T. Up-regulation of miR-224 promotes cancer cell proliferation and invasion and predicts relapse of colorectal cancer. Cancer Cell. Int. 2013, 13, 104. [Google Scholar] [CrossRef] [Green Version]
- Fang, X.; Dong, Y.; Yang, R.; Wei, L. LINC00619 restricts gastric cancer progression by preventing microRNA-224-5p-mediated inhibition of OPCML. Arch. Biochem. Biophys. 2020, 689, 108390. [Google Scholar] [CrossRef]
- Pfeffer, S.R.; Grossmann, K.F.; Cassidy, P.B.; Yang, C.H.; Fan, M.; Kopelovich, L.; Leachman, S.A.; Pfeffer, L.M. Detection of Exosomal miRNAs in the Plasma of Melanoma Patients. J. Clin. Med. 2015, 4, 2012–2027. [Google Scholar] [CrossRef]
- Zhang, J.; Hou, L.; Liang, R.; Chen, X.; Zhang, R.; Chen, W.; Zhu, J. CircDLST promotes the tumorigenesis and metastasis of gastric cancer by sponging miR-502-5p and activating the NRAS/MEK1/ERK1/2 signaling. Mol. Cancer 2019, 18, 80. [Google Scholar] [CrossRef]
- Yan, B.; Guo, Q.; Fu, F.-J.; Wang, Z.; Yin, Z.; Wei, Y.-B.; Yang, J.-R. The role of miR-29b in cancer: Regulation, function, and signaling. Oncol. Targ. Ther. 2015, 8, 539–548. [Google Scholar]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.-L.; Bao, H.-G.; Zheng, C.-L.; Teng, C.; Bai, M.-H. MiR-130a regulating the biological function of colon cancer by targeting inhibition of PTEN. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1786–1793. [Google Scholar] [PubMed]
- Redmer, T. Deciphering mechanisms of brain metastasis in melanoma-the gist of the matter. Mol. Cancer 2018, 17, 106. [Google Scholar] [CrossRef] [PubMed]
- Tentori, L.; Leonetti, C.; Scarsella, M.; D’Amati, G.; Vergati, M.; Portarena, I.; Xu, W.; Kalish, V.; Zupi, G.; Zhang, J.; et al. Systemic administration of GPI 15427, a novel poly(ADP-ribose) polymerase-1 inhibitor, increases the antitumor activity of temozolomide against intracranial melanoma, glioma, lymphoma. Clin. Cancer Res. 2003, 9, 5370–5379. [Google Scholar]
- Palmieri, V.; Lucchetti, D.; Gatto, I.; Maiorana, A.; Marcantoni, M.; Maulucci, G.; Papi, M.; Pola, R.; De Spirito, M.; Sgambato, A. Dynamic light scattering for the characterization and counting of extracellular vesicles: A powerful noninvasive tool. J. Nanop. Res. 2014, 16, 2583. [Google Scholar] [CrossRef]
- Papi, M.; Lauriola, M.C.; Palmieri, V.; Ciasca, G.; Maulucci, G.; De Spirito, M. Plasma protein corona reduces the haemolytic activity of graphene oxide nano and micro flakes. RSC Adv. 2015, 5, 81638–81641. [Google Scholar] [CrossRef]
- Kirschner, M.B.; Kao, S.C.; Edelman, J.J.; Armstrong, N.J.; Vallely, M.P.; van Zandwijk, N.; Reid, G. Haemolysis during sample preparation alters microRNA content of plasma. PLoS ONE 2011, 6, e24145. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Kroh, E.; Wood, B.; Arroyo, J.D.; Dougherty, K.J.; Miyaji, M.M.; Tait, J.F.; Tewari, M. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev. Res. 2012, 5, 492–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vergoulis, T.; Vlachos, I.S.; Alexiou, P.; Georgakilas, G.; Maragkakis, M.; Reczko, M.; Gerangelos, S.; Koziris, N.; Dalamagas, T.; Hatzigeorgiou, A.G. TarBase 6.0: Capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res. 2012, 40, D222–D229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.; Kim, J.; Jha, S.; Aboul-ela, F. A mechanism for S-adenosyl methionine assisted formation of a riboswitch conformation: A small molecule with a strong arm. Nucleic Acids Res. 2009, 37, 6528–6539. [Google Scholar] [CrossRef]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 18, 13. [Google Scholar] [CrossRef] [PubMed]







© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guglielmi, L.; Nardella, M.; Musa, C.; Cifola, I.; Porru, M.; Cardinali, B.; Iannetti, I.; Di Pietro, C.; Bolasco, G.; Palmieri, V.; et al. Circulating miRNAs in Small Extracellular Vesicles Secreted by a Human Melanoma Xenograft in Mouse Brains. Cancers 2020, 12, 1635. https://doi.org/10.3390/cancers12061635
Guglielmi L, Nardella M, Musa C, Cifola I, Porru M, Cardinali B, Iannetti I, Di Pietro C, Bolasco G, Palmieri V, et al. Circulating miRNAs in Small Extracellular Vesicles Secreted by a Human Melanoma Xenograft in Mouse Brains. Cancers. 2020; 12(6):1635. https://doi.org/10.3390/cancers12061635
Chicago/Turabian StyleGuglielmi, Loredana, Marta Nardella, Carla Musa, Ingrid Cifola, Manuela Porru, Beatrice Cardinali, Ilaria Iannetti, Chiara Di Pietro, Giulia Bolasco, Valentina Palmieri, and et al. 2020. "Circulating miRNAs in Small Extracellular Vesicles Secreted by a Human Melanoma Xenograft in Mouse Brains" Cancers 12, no. 6: 1635. https://doi.org/10.3390/cancers12061635
APA StyleGuglielmi, L., Nardella, M., Musa, C., Cifola, I., Porru, M., Cardinali, B., Iannetti, I., Di Pietro, C., Bolasco, G., Palmieri, V., Vilardo, L., Panini, N., Bonaventura, F., Papi, M., Scavizzi, F., Raspa, M., Leonetti, C., Falcone, G., Felsani, A., & D’Agnano, I. (2020). Circulating miRNAs in Small Extracellular Vesicles Secreted by a Human Melanoma Xenograft in Mouse Brains. Cancers, 12(6), 1635. https://doi.org/10.3390/cancers12061635