Downregulation of the Ubiquitin-E3 Ligase RNF123 Promotes Upregulation of the NF-κB1 Target SerpinE1 in Aggressive Glioblastoma Tumors
Abstract
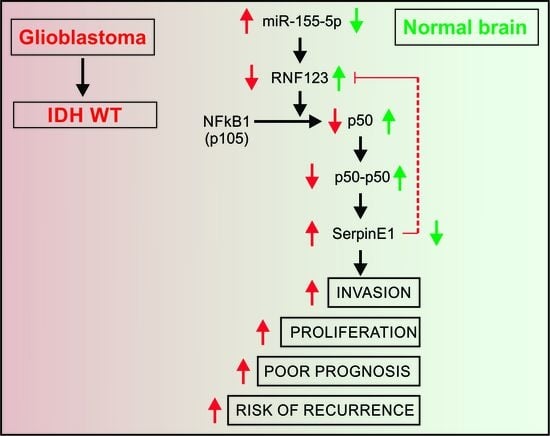
:1. Introduction
2. Results
2.1. RNF123 Downregulation is Associated with a Poor Prognosis in GB
2.2. P50 Protein Levels but Not p27 Are Associated with RNF123 Expression in GB
2.3. RNF123 Overexpression Suppresses Cell Proliferation and Invasion
2.4. Expression of SerpinE1 Is Negatively Associated with RNF123 Expression
2.5. SerpinE1 Expression Is Associated with a Poor Prognosis in GB
2.6. SerpinE1-OV Promotes Proliferation, Invasion, and Colony-Formation Ability of GB Cells
2.7. SerpinE1 Overexpression Rescued the Inhibitory Effects Mediated by RNF123-OV- in GB Cell Lines
2.8. Concurrent Low RNF123 and High SerpinE1 Expression Is Associated with a Poor Prognosis
2.9. MiR-155 Suppresses RNF123 Expression in GB and Is Associated with a Poor Prognosis
3. Discussion
4. Materials and Methods
4.1. Ethics Approval and Consent to Participate
4.2. Clinical Characteristics of Patients
4.3. Cell Lines
4.4. Reverse-Phase Protein Array Analysis
4.5. Immunohistochemistry for Tissue Microarrays
4.6. Stable Clones Overexpressing RNF123
4.7. Stable Clones Overexpressing SERPINE1
4.8. Bioinformatics Analysis
4.9. Data Availability
4.10. Information Available in Supplementary Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 3′-UTR | 3′-Untranslated Region |
| Abs | Antibodies |
| B2MG | Beta-2-Microglobulin |
| CI | Confidence Interval |
| CIF | Cumulative Incidence Function |
| EV | Empty Vector 1 and 2 |
| HR | Hazard Risk |
| FFPE | Formalin-Fixed Paraffin Embedded |
| GB | Glioblastoma |
| IDH | Isocitrate Dehydrogenase 1 and 2 Gene |
| IHC | Immunohistochemistry |
| KPS | Karnofsky Performance Score |
| NF-κB | Nuclear Factor kappa-Light-Chain-Enhancer of Activated B Cells |
| NS | Non-Significant |
| MiR | MicroRNA |
| MGMT | Methyl guanine methyltransferase |
| OS | Overall Survival |
| PBS | Phosphate Buffer Saline |
| PFS | Progression-Free Survival |
| RPPA | Reverse-Phase Protein Arrays |
| RR | Relative Risk |
| RT-qPCR | Real-time quantitative polymerase chain reaction |
| SD | Standard Deviation |
| TCGA | The Cancer Genome Atlas Project |
| TMA | Tissue Microarray |
| UPS | Ubiquitin-proteasome system |
| WT | Wild Type |
| WHO | World Health Organization |
References
- Lukas, R.V.; Wainwright, D.A.; Ladomersky, E.; Sachdev, S.; Sonabend, A.M.; Stupp, R. Newly Diagnosed Glioblastoma: A Review on Clinical Management. Oncology 2019, 33, 91–100. [Google Scholar] [PubMed]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, E.; Yong, R.L.; Paddison, P.; Zhu, J. Comparison of glioblastoma (GBM) molecular classification methods. Semin. Cancer Biol. 2018, 53, 201–211. [Google Scholar] [CrossRef]
- Taylor, O.G.; Brzozowski, J.S.; Skelding, K.A. Glioblastoma Multiforme: An Overview of Emerging Therapeutic Targets. Front. Oncol. 2019, 9, 963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Hu, B.; Hu, X.; Kim, H.; Squatrito, M.; Scarpace, L.; deCarvalho, A.C.; Lyu, S.; Li, P.; Li, Y.; et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017, 32, 42–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wesseling, P.; Capper, D. WHO 2016 Classification of gliomas. Neuropathol. Appl. Neurobiol. 2018, 44, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Sanson, M.; Marie, Y.; Paris, S.; Idbaih, A.; Laffaire, J.; Ducray, F.; El Hallani, S.; Boisselier, B.; Mokhtari, K.; Hoang-Xuan, K.; et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J. Clin. Oncol. 2009, 27, 4150–4154. [Google Scholar] [CrossRef]
- Kumari, N.; Jaynes, P.W.; Saei, A.; Iyengar, P.V.; Richard, J.L.C.; Eichhorn, P.J.A. The roles of ubiquitin modifying enzymes in neoplastic disease. Biochim. Biophys. Acta Rev. Cancer 2017, 1868, 456–483. [Google Scholar] [CrossRef]
- Iida, Y.; Ciechanover, A.; Marzese, D.M.; Hata, K.; Bustos, M.; Ono, S.; Wang, J.; Salomon, M.P.; Tran, K.; Lam, S.; et al. Epigenetic Regulation of KPC1 Ubiquitin Ligase Affects the NF-kappaB Pathway in Melanoma. Clin. Cancer Res. 2017, 23, 4831–4842. [Google Scholar] [CrossRef] [Green Version]
- Ciechanover, A. The unravelling of the ubiquitin system. Nat. Rev. Mol. Cell Biol. 2015, 16, 322–324. [Google Scholar] [CrossRef]
- Kravtsova-Ivantsiv, Y.; Ciechanover, A. The ubiquitin-proteasome system and activation of NF-kappaB: Involvement of the ubiquitin ligase KPC1 in p105 processing and tumor suppression. Mol. Cell. Oncol. 2015, 2, e1054552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Ma, L.; Wang, B.; Liu, J.; Wei, W. E3 ubiquitin ligases in cancer and implications for therapies. Cancer Metastasis Rev. 2017, 36, 683–702. [Google Scholar] [CrossRef] [PubMed]
- Perkins, N.D. The diverse and complex roles of NF-kappaB subunits in cancer. Nat. Rev. Cancer 2012, 12, 121–132. [Google Scholar] [CrossRef] [PubMed]
- DiDonato, J.A.; Mercurio, F.; Karin, M. NF-kappaB and the link between inflammation and cancer. Immunol. Rev. 2012, 246, 379–400. [Google Scholar] [CrossRef]
- Gray, G.K.; McFarland, B.C.; Nozell, S.E.; Benveniste, E.N. NF-kappaB and STAT3 in glioblastoma: Therapeutic targets coming of age. Expert Rev. Neurother. 2014, 14, 1293–1306. [Google Scholar] [CrossRef] [Green Version]
- Cartwright, T.; Perkins, N.D.; C, L.W. NFKB1: A suppressor of inflammation, ageing and cancer. FEBS J. 2016, 283, 1812–1822. [Google Scholar] [CrossRef] [Green Version]
- Kamura, T.; Hara, T.; Matsumoto, M.; Ishida, N.; Okumura, F.; Hatakeyama, S.; Yoshida, M.; Nakayama, K.; Nakayama, K.I. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat. Cell Biol. 2004, 6, 1229–1235. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, S.; Wu, X.; Huan, W.; Liu, Z.; Wei, H.; Shen, A.; Teng, H. KPC1 expression and essential role after acute spinal cord injury in adult rat. Neurochem. Res. 2011, 36, 549–558. [Google Scholar] [CrossRef]
- Kravtsova-Ivantsiv, Y.; Shomer, I.; Cohen-Kaplan, V.; Snijder, B.; Superti-Furga, G.; Gonen, H.; Sommer, T.; Ziv, T.; Admon, A.; Naroditsky, I.; et al. KPC1-mediated ubiquitination and proteasomal processing of NF-kappaB1 p105 to p50 restricts tumor growth. Cell 2015, 161, 333–347. [Google Scholar] [CrossRef] [Green Version]
- Hou, B.; Eren, M.; Painter, C.A.; Covington, J.W.; Dixon, J.D.; Schoenhard, J.A.; Vaughan, D.E. Tumor necrosis factor alpha activates the human plasminogen activator inhibitor-1 gene through a distal nuclear factor kappaB site. J. Biol. Chem. 2004, 279, 18127–18136. [Google Scholar] [CrossRef] [Green Version]
- Kwaan, H.C.; Mazar, A.P.; McMahon, B.J. The apparent uPA/PAI-1 paradox in cancer: More than meets the eye. Semin. Thromb. Hemost. 2013, 39, 382–391. [Google Scholar] [CrossRef]
- Nakatsuka, E.; Sawada, K.; Nakamura, K.; Yoshimura, A.; Kinose, Y.; Kodama, M.; Hashimoto, K.; Mabuchi, S.; Makino, H.; Morii, E.; et al. Plasminogen activator inhibitor-1 is an independent prognostic factor of ovarian cancer and IMD-4482, a novel plasminogen activator inhibitor-1 inhibitor, inhibits ovarian cancer peritoneal dissemination. Oncotarget 2017, 8, 89887–89902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavon, M.A.; Arroyo-Solera, I.; Tellez-Gabriel, M.; Leon, X.; Viros, D.; Lopez, M.; Gallardo, A.; Cespedes, M.V.; Casanova, I.; Lopez-Pousa, A.; et al. Enhanced cell migration and apoptosis resistance may underlie the association between high SERPINE1 expression and poor outcome in head and neck carcinoma patients. Oncotarget 2015, 6, 29016–29033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Placencio, V.R.; DeClerck, Y.A. Plasminogen Activator Inhibitor-1 in Cancer: Rationale and Insight for Future Therapeutic Testing. Cancer Res. 2015, 75, 2969–2974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Placencio, V.R.; Ichimura, A.; Miyata, T.; DeClerck, Y.A. Small Molecule Inhibitors of Plasminogen Activator Inhibitor-1 Elicit Anti-Tumorigenic and Anti-Angiogenic Activity. PLoS ONE 2015, 10, e0133786. [Google Scholar] [CrossRef] [Green Version]
- Bryan, L.; Paugh, B.S.; Kapitonov, D.; Wilczynska, K.M.; Alvarez, S.M.; Singh, S.K.; Milstien, S.; Spiegel, S.; Kordula, T. Sphingosine-1-phosphate and interleukin-1 independently regulate plasminogen activator inhibitor-1 and urokinase-type plasminogen activator receptor expression in glioblastoma cells: Implications for invasiveness. Mol. Cancer Res. MCR 2008, 6, 1469–1477. [Google Scholar] [CrossRef] [Green Version]
- Look, M.P.; van Putten, W.L.; Duffy, M.J.; Harbeck, N.; Christensen, I.J.; Thomssen, C.; Kates, R.; Spyratos, F.; Ferno, M.; Eppenberger-Castori, S.; et al. Pooled analysis of prognostic impact of urokinase-type plasminogen activator and its inhibitor PAI-1 in 8377 breast cancer patients. J. Natl. Cancer Inst. 2002, 94, 116–128. [Google Scholar] [CrossRef] [Green Version]
- Spyratos, F.; Bouchet, C.; Tozlu, S.; Labroquere, M.; Vignaud, S.; Becette, V.; Lidereau, R.; Bieche, I. Prognostic value of uPA, PAI-1 and PAI-2 mRNA expression in primary breast cancer. Anticancer Res. 2002, 22, 2997–3003. [Google Scholar]
- Hildenbrand, R.; Arens, N. Protein and mRNA expression of uPAR and PAI-1 in myoepithelial cells of early breast cancer lesions and normal breast tissue. Br. J. Cancer 2004, 91, 564–571. [Google Scholar] [CrossRef] [Green Version]
- Fredstorp-Lidebring, M.; Bendahl, P.O.; Brunner, N.; Casslen, B.; Hogberg, T.; Langstrom-Einarsson, E.; Willen, R.; Ferno, M. Urokinase plasminogen activator and its inhibitor, PAI-1, in association with progression-free survival in early stage endometrial cancer. Eur. J. Cancer 2001, 37, 2339–2348. [Google Scholar] [CrossRef]
- Nekarda, H.; Schmitt, M.; Ulm, K.; Wenninger, A.; Vogelsang, H.; Becker, K.; Roder, J.D.; Fink, U.; Siewert, J.R. Prognostic impact of urokinase-type plasminogen activator and its inhibitor PAI-1 in completely resected gastric cancer. Cancer Res. 1994, 54, 2900–2907. [Google Scholar] [PubMed]
- Papadopoulou, S.; Scorilas, A.; Yotis, J.; Arnogianaki, N.; Plataniotis, G.; Agnanti, N.; Talieri, M. Significance of urokinase-type plasminogen activator and plasminogen activator inhibitor-1 (PAI-1) expression in human colorectal carcinomas. Tumour Biol. 2002, 23, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Werle, B.; Kotzsch, M.; Lah, T.T.; Kos, J.; Gabrijelcic-Geiger, D.; Spiess, E.; Schirren, J.; Ebert, W.; Fiehn, W.; Luther, T.; et al. Cathepsin B, plasminogenactivator-inhibitor (PAI-1) and plasminogenactivator-receptor (uPAR) are prognostic factors for patients with non-small cell lung cancer. Anticancer Res. 2004, 24, 4147–4161. [Google Scholar] [PubMed]
- Zubac, D.P.; Wentzel-Larsen, T.; Seidal, T.; Bostad, L. Type 1 plasminogen activator inhibitor (PAI-1) in clear cell renal cell carcinoma (CCRCC) and its impact on angiogenesis, progression and patient survival after radical nephrectomy. BMC Urol. 2010, 10, 20. [Google Scholar] [CrossRef] [Green Version]
- Colin, C.; Voutsinos-Porche, B.; Nanni, I.; Fina, F.; Metellus, P.; Intagliata, D.; Baeza, N.; Bouvier, C.; Delfino, C.; Loundou, A.; et al. High expression of cathepsin B and plasminogen activator inhibitor type-1 are strong predictors of survival in glioblastomas. Acta Neuropathol. 2009, 118, 745–754. [Google Scholar] [CrossRef]
- Kit, O.I.; Frantsiyants, E.M.; Kozlova, L.S.; Rostorguev, E.E.; Balyazin-Parfenov, I.V.; Pogorelova, Y.A. A plasminogen regulation system in brain tumors. Zhurnal Vopr. Neirokhirurgii Im. N. N. Burdenko 2017, 81, 22–27. [Google Scholar] [CrossRef]
- Iwadate, Y.; Hayama, M.; Adachi, A.; Matsutani, T.; Nagai, Y.; Hiwasa, T.; Saeki, N. High serum level of plasminogen activator inhibitor-1 predicts histological grade of intracerebral gliomas. Anticancer Res. 2008, 28, 415–418. [Google Scholar]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Hidalgo, B.; Goodman, M. Multivariate or multivariable regression? Am. J. Public Health 2013, 103, 39–40. [Google Scholar] [CrossRef]
- Xu, L.; Liu, F.; Zhu, J.; Zhu, L.; Jiang, F. Expression and Role of KIP1 Ubiquitylation-Promoting Complex 2 in Astrocytes after Spinal Cord Injury in RATS. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi = Zhongguo Xiufu Chongjian Waike Zazhi = Chin. J. Reparative Reconstr. Surg. 2015, 29, 978–985. [Google Scholar]
- Wu, X.; Wang, Y.; Yu, T.; Nie, E.; Hu, Q.; Wu, W.; Zhi, T.; Jiang, K.; Wang, X.; Lu, X.; et al. Blocking MIR155HG/miR-155 axis inhibits mesenchymal transition in glioma. Neuro-Oncology 2017, 19, 1195–1205. [Google Scholar] [CrossRef]
- Liu, Q.; Zou, R.; Zhou, R.; Gong, C.; Wang, Z.; Cai, T.; Tan, C.; Fang, J. miR-155 Regulates Glioma Cells Invasion and Chemosensitivity by p38 Isforms In Vitro. J. Cell. Biochem. 2015, 116, 1213–1221. [Google Scholar] [CrossRef]
- Iida, Y.; Salomon, M.P.; Hata, K.; Tran, K.; Ohe, S.; Griffiths, C.F.; Hsu, S.C.; Nelson, N.; Hoon, D.S.B. Predominance of triple wild-type and IGF2R mutations in mucosal melanomas. BMC Cancer 2018, 18, 1054. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Adegoke, O.A.; Nepveu, A.; Nakayama, K.I.; Bedard, N.; Cheng, D.; Peng, J.; Wing, S.S. USP19 deubiquitinating enzyme supports cell proliferation by stabilizing KPC1, a ubiquitin ligase for p27Kip1. Mol. Cell. Boil. 2009, 29, 547–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Xie, W.; Kuhn, D.J.; Voorhees, P.M.; Lopez-Girona, A.; Mendy, D.; Corral, L.G.; Krenitsky, V.P.; Xu, W.; Moutouh-de Parseval, L.; et al. Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood 2008, 111, 4690–4699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitcomb, E.A.; Tsai, Y.C.; Basappa, J.; Liu, K.; Le Feuvre, A.K.; Weissman, A.M.; Taylor, A. Stabilization of p27(Kip1)/CDKN1B by UBCH7/UBE2L3 catalyzed ubiquitinylation: A new paradigm in cell-cycle control. FASEB J. 2019, 33, 1235–1247. [Google Scholar] [CrossRef] [PubMed]
- Cahill, K.E.; Morshed, R.A.; Yamini, B. Nuclear factor-kappaB in glioblastoma: Insights into regulators and targeted therapy. Neuro-Oncology 2016, 18, 329–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behnan, J.; Finocchiaro, G.; Hanna, G. The landscape of the mesenchymal signature in brain tumours. Brain 2019, 142, 847–866. [Google Scholar] [CrossRef] [Green Version]
- Patil, V.; Mahalingam, K. Comprehensive analysis of Reverse Phase Protein Array data reveals characteristic unique proteomic signatures for glioblastoma subtypes. Gene 2018, 685, 85–95. [Google Scholar] [CrossRef]
- Lin, S.Y.; Huang, S.K.; Huynh, K.T.; Salomon, M.P.; Chang, S.-C.; Marzese, D.M.; Lanman, R.B.; Talasaz, A.; Hoon, D.S.B. Multiplex Gene Profiling of Cell-Free DNA in Patients With Metastatic Melanoma for Monitoring Disease. JCO Precis. Oncol. 2018, 2, 1–30. [Google Scholar] [CrossRef]
- Akbani, R.; Ng, P.K.; Werner, H.M.; Shahmoradgoli, M.; Zhang, F.; Ju, Z.; Liu, W.; Yang, J.Y.; Yoshihara, K.; Li, J.; et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat. Commun. 2014, 5, 3887. [Google Scholar] [CrossRef] [PubMed]







© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Bustos, M.A.; Zhang, X.; Ramos, R.I.; Tan, C.; Iida, Y.; Chang, S.-C.; Salomon, M.P.; Tran, K.; Gentry, R.; et al. Downregulation of the Ubiquitin-E3 Ligase RNF123 Promotes Upregulation of the NF-κB1 Target SerpinE1 in Aggressive Glioblastoma Tumors. Cancers 2020, 12, 1081. https://doi.org/10.3390/cancers12051081
Wang X, Bustos MA, Zhang X, Ramos RI, Tan C, Iida Y, Chang S-C, Salomon MP, Tran K, Gentry R, et al. Downregulation of the Ubiquitin-E3 Ligase RNF123 Promotes Upregulation of the NF-κB1 Target SerpinE1 in Aggressive Glioblastoma Tumors. Cancers. 2020; 12(5):1081. https://doi.org/10.3390/cancers12051081
Chicago/Turabian StyleWang, Xiaowen, Matias A. Bustos, Xiaoqing Zhang, Romela Irene Ramos, Cong Tan, Yuuki Iida, Shu-Ching Chang, Matthew P. Salomon, Kevin Tran, Rebecca Gentry, and et al. 2020. "Downregulation of the Ubiquitin-E3 Ligase RNF123 Promotes Upregulation of the NF-κB1 Target SerpinE1 in Aggressive Glioblastoma Tumors" Cancers 12, no. 5: 1081. https://doi.org/10.3390/cancers12051081