An Amplified Fatty Acid-Binding Protein Gene Cluster in Prostate Cancer: Emerging Roles in Lipid Metabolism and Metastasis
Abstract
:Simple Summary
Abstract
1. Introduction
2. A FABP Gene Cluster Is Preferentially Amplified and Overexpressed in Metastatic PCa and Associated with Poor Clinical Outcomes
3. Roles of FABPs in PCa Metastatic Progression
3.1. FABPs in Fatty Acid Uptake and Lipid Droplet Formation
3.2. FABPs Modulate Lipid Metabolism That Fuels PCa Cell Dissemination and Metastasis
3.3. FABPs Induce Epithelial-to-Mesenchymal Transition to Prime PCa Cells for Metastasis
3.4. FABPs Promote Cancer Cell Motility and Invasion
3.5. FABPs Promotes PCa Metastasis by Stimulating Angiogenesis
4. The Molecular Pathways Underlying FABP-Induced PCa Metastasis
5. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massague, J.; Obenauf, A.C. Metastatic colonization by circulating tumour cells. Nature 2016, 529, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Talmadge, J.E.; Fidler, I.J. AACR centennial series: The biology of cancer metastasis: Historical perspective. Cancer Res. 2010, 70, 5649–5669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asmane, I.; Ceraline, J.; Duclos, B.; Rob, L.; Litique, V.; Barthelemy, P.; Bergerat, J.P.; Dufour, P.; Kurtz, J.E. New strategies for medical management of castration-resistant prostate cancer. Oncology (Williston Park) 2011, 80, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gandaglia, G.; Abdollah, F.; Schiffmann, J.; Trudeau, V.; Shariat, S.F.; Kim, S.P.; Perrotte, P.; Montorsi, F.; Briganti, A.; Trinh, Q.D.; et al. Distribution of metastatic sites in patients with prostate cancer: A population-based analysis. Prostate 2014, 74, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Shou, J.; Zhang, Q.; Wang, S.; Zhang, D. The prognosis of different distant metastases pattern in prostate cancer: A population based retrospective study. Prostate 2018, 78, 491–497. [Google Scholar] [CrossRef]
- Clarke, N.W.; Hart, C.A.; Brown, M.D. Molecular mechanisms of metastasis in prostate cancer. Asian J. Androl. 2009, 11, 57–67. [Google Scholar] [CrossRef] [Green Version]
- Weidle, U.H.; Birzele, F.; Kollmorgen, G.; Ruger, R. Molecular Mechanisms of Bone Metastasis. Cancer Genom. Proteom. 2016, 13, 1–12. [Google Scholar]
- Chiang, A.C.; Massague, J. Molecular basis of metastasis. N. Engl. J. Med. 2008, 359, 2814–2823. [Google Scholar] [CrossRef] [Green Version]
- Gleason, D.F. Classification of prostatic carcinomas. Cancer Chemother. Rep. 1966, 50, 125–128. [Google Scholar]
- Gleason, D.F.; Mellinger, G.T. Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. J. Urol. 1974, 111, 58–64. [Google Scholar] [CrossRef]
- Catalona, W.J.; Richie, J.P.; Ahmann, F.R.; Hudson, M.A.; Scardino, P.T.; Flanigan, R.C.; deKernion, J.B.; Ratliff, T.L.; Kavoussi, L.R.; Dalkin, B.L.; et al. Comparison of digital rectal examination and serum prostate specific antigen in the early detection of prostate cancer: Results of a multicenter clinical trial of 6630 men. J. Urol. 1994, 151, 1283–1290. [Google Scholar] [CrossRef]
- Draisma, G.; Etzioni, R.; Tsodikov, A.; Mariotto, A.; Wever, E.; Gulati, R.; Feuer, E.; de Koning, H. Lead time and overdiagnosis in prostate-specific antigen screening: Importance of methods and context. J. Natl. Cancer Inst. 2009, 101, 374–383. [Google Scholar] [CrossRef] [Green Version]
- Ramirez, M.L.; Nelson, E.C.; Evans, C.P. Beyond prostate-specific antigen: Alternate serum markers. Prostate Cancer Prostatic Dis. 2008, 11, 216–229. [Google Scholar] [CrossRef] [Green Version]
- Andersen, K.F.; Divilov, V.; Sevak, K.; Koziorowski, J.; Lewis, J.S.; Pillarsetty, N. Influence of free fatty acids on glucose uptake in prostate cancer cells. Nucl. Med. Biol. 2014, 41, 254–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y. Fatty acid oxidation is a dominant bioenergetic pathway in prostate cancer. Prostate Cancer Prostat. Dis. 2006, 9, 230–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zuckier, L.S.; Ghesani, N.V. Dominant uptake of fatty acid over glucose by prostate cells: A potential new diagnostic and therapeutic approach. Anticancer Res. 2010, 30, 369–374. [Google Scholar] [PubMed]
- Haunerland, N.H.; Spener, F. Fatty acid-binding proteins-insights from genetic manipulations. Prog. Lipid Res. 2004, 43, 328–349. [Google Scholar] [CrossRef]
- Chmurzynska, A. The multigene family of fatty acid-binding proteins (FABPs): Function, structure and polymorphism. J. Appl. Genet. 2006, 47, 39–48. [Google Scholar] [CrossRef]
- Liu, R.Z.; Li, X.; Godbout, R. A novel fatty acid-binding protein (FABP) gene resulting from tandem gene duplication in mammals: Transcription in rat retina and testis. Genomics 2008, 92, 436–445. [Google Scholar] [CrossRef] [Green Version]
- Elsherbiny, M.E.; Emara, M.; Godbout, R. Interaction of brain fatty acid-binding protein with the polyunsaturated fatty acid environment as a potential determinant of poor prognosis in malignant glioma. Prog. Lipid Res. 2013, 52, 562–570. [Google Scholar] [CrossRef] [Green Version]
- Storch, J.; Corsico, B. The emerging functions and mechanisms of mammalian fatty acid-binding proteins. Annu. Rev. Nutr. 2008, 28, 73–95. [Google Scholar] [CrossRef] [PubMed]
- Amiri, M.; Yousefnia, S.; Seyed Forootan, F.; Peymani, M.; Ghaedi, K.; Nasr Esfahani, M.H. Diverse roles of fatty acid binding proteins (FABPs) in development and pathogenesis of cancers. Gene 2018, 676, 171–183. [Google Scholar] [CrossRef]
- Gharpure, K.M.; Pradeep, S.; Sans, M.; Rupaimoole, R.; Ivan, C.; Wu, S.Y.; Bayraktar, E.; Nagaraja, A.S.; Mangala, L.S.; Zhang, X.; et al. FABP4 as a key determinant of metastatic potential of ovarian cancer. Nat. Commun. 2018, 9, 2923. [Google Scholar] [CrossRef] [Green Version]
- Tian, W.; Zhang, W.; Zhang, Y.; Zhu, T.; Hua, Y.; Li, H.; Zhang, Q.; Xia, M. FABP4 promotes invasion and metastasis of colon cancer by regulating fatty acid transport. Cancer Cell Int. 2020, 20, 512. [Google Scholar] [CrossRef] [PubMed]
- Uehara, H.; Takahashi, T.; Oha, M.; Ogawa, H.; Izumi, K. Exogenous fatty acid binding protein 4 promotes human prostate cancer cell progression. Int. J. Cancer 2014, 135, 2558–2568. [Google Scholar] [CrossRef]
- Adamson, J.; Morgan, E.A.; Beesley, C.; Mei, Y.; Foster, C.S.; Fujii, H.; Rudland, P.S.; Smith, P.H.; Ke, Y. High-level expression of cutaneous fatty acid-binding protein in prostatic carcinomas and its effect on tumorigenicity. Oncogene 2003, 22, 2739–2749. [Google Scholar] [CrossRef] [Green Version]
- Forootan, S.S.; Bao, Z.Z.; Forootan, F.S.; Kamalian, L.; Zhang, Y.; Bee, A.; Foster, C.S.; Ke, Y. Atelocollagen-delivered siRNA targeting the FABP5 gene as an experimental therapy for prostate cancer in mouse xenografts. Int. J. Oncol. 2010, 36, 69–76. [Google Scholar]
- Liu, R.Z.; Choi, W.S.; Jain, S.; Dinakaran, D.; Xu, X.; Han, W.H.; Yang, X.H.; Glubrecht, D.D.; Moore, R.B.; Lemieux, H.; et al. The FABP12/PPARgamma pathway promotes metastatic transformation by inducing epithelial-to-mesenchymal transition and lipid-derived energy production in prostate cancer cells. Mol. Oncol. 2020, 14, 3100–3120. [Google Scholar] [CrossRef]
- Thumser, A.E.; Moore, J.B.; Plant, N.J. Fatty acid binding proteins: Tissue-specific functions in health and disease. Curr. Opin. Clin. Nutr. Metab. Care 2014, 17, 124–129. [Google Scholar] [CrossRef] [Green Version]
- Furuhashi, M.; Hotamisligil, G.S. Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 2008, 7, 489–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hotamisligil, G.S.; Bernlohr, D.A. Metabolic functions of FABPs--mechanisms and therapeutic implications. Nat. Rev. Endocrinol. 2015, 11, 592–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schug, T.T.; Berry, D.C.; Shaw, N.S.; Travis, S.N.; Noy, N. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell 2007, 129, 723–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schug, T.T.; Berry, D.C.; Toshkov, I.A.; Cheng, L.; Nikitin, A.Y.; Noy, N. Overcoming retinoic acid-resistance of mammary carcinomas by diverting retinoic acid from PPARbeta/delta to RAR. Proc. Natl. Acad. Sci. USA 2008, 105, 7546–7551. [Google Scholar] [CrossRef] [Green Version]
- Bao, Z.; Malki, M.I.; Forootan, S.S.; Adamson, J.; Forootan, F.S.; Chen, D.; Foster, C.S.; Rudland, P.S.; Ke, Y. A novel cutaneous Fatty Acid-binding protein-related signaling pathway leading to malignant progression in prostate cancer cells. Genes Cancer 2013, 4, 297–314. [Google Scholar] [CrossRef]
- Forootan, F.S.; Forootan, S.S.; Malki, M.I.; Chen, D.; Li, G.; Lin, K.; Rudland, P.S.; Foster, C.S.; Ke, Y. The expression of C-FABP and PPARgamma and their prognostic significance in prostate cancer. Int. J. Oncol. 2014, 44, 265–275. [Google Scholar] [CrossRef]
- Morgan, E.; Kannan-Thulasiraman, P.; Noy, N. Involvement of Fatty Acid Binding Protein 5 and PPARbeta/delta in Prostate Cancer Cell Growth. PPAR Res. 2010, 2010. [Google Scholar] [CrossRef] [Green Version]
- Schroeder, F.; Petrescu, A.D.; Huang, H.; Atshaves, B.P.; McIntosh, A.L.; Martin, G.G.; Hostetler, H.A.; Vespa, A.; Landrock, D.; Landrock, K.K.; et al. Role of fatty acid binding proteins and long chain fatty acids in modulating nuclear receptors and gene transcription. Lipids 2008, 43, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Cher, M.L.; Bova, G.S.; Moore, D.H.; Small, E.J.; Carroll, P.R.; Pin, S.S.; Epstein, J.I.; Isaacs, W.B.; Jensen, R.H. Genetic alterations in untreated metastases and androgen-independent prostate cancer detected by comparative genomic hybridization and allelotyping. Cancer Res. 1996, 56, 3091–3102. [Google Scholar]
- Santarius, T.; Shipley, J.; Brewer, D.; Stratton, M.R.; Cooper, C.S. A census of amplified and overexpressed human cancer genes. Nat. Rev. Cancer 2010, 10, 59–64. [Google Scholar] [CrossRef]
- Porkka, K.; Saramaki, O.; Tanner, M.; Visakorpi, T. Amplification and overexpression of Elongin C gene discovered in prostate cancer by cDNA microarrays. Lab. Investig. J. Tech. Methods Pathol. 2002, 82, 629–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubin, M.A.; Varambally, S.; Beroukhim, R.; Tomlins, S.A.; Rhodes, D.R.; Paris, P.L.; Hofer, M.D.; Storz-Schweizer, M.; Kuefer, R.; Fletcher, J.A.; et al. Overexpression, amplification, and androgen regulation of TPD52 in prostate cancer. Cancer Res. 2004, 64, 3814–3822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Xu, J.; Saramaki, O.; Visakorpi, T.; Sutherland, W.M.; Zhou, J.; Sen, B.; Lim, S.D.; Mabjeesh, N.; Amin, M.; et al. PrLZ, a novel prostate-specific and androgen-responsive gene of the TPD52 family, amplified in chromosome 8q21.1 and overexpressed in human prostate cancer. Cancer Res. 2004, 64, 1589–1594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, B.S.; Schultz, N.; Hieronymus, H.; Gopalan, A.; Xiao, Y.; Carver, B.S.; Arora, V.K.; Kaushik, P.; Cerami, E.; Reva, B.; et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010, 18, 11–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawaguchi, K.; Kinameri, A.; Suzuki, S.; Senga, S.; Ke, Y.; Fujii, H. The Cancer-Promoting Gene Fatty Acid-Binding Protein 5 (FABP5) is Epigenetically Regulated During Human Prostate Carcinogenesis. Biochem. J. 2015. [Google Scholar] [CrossRef] [PubMed]
- Al-Jameel, W.; Gou, X.; Forootan, S.S.; Al Fayi, M.S.; Rudland, P.S.; Forootan, F.S.; Zhang, J.; Cornford, P.A.; Hussain, S.A.; Ke, Y. Inhibitor SBFI26 suppresses the malignant progression of castration-resistant PC3-M cells by competitively binding to oncogenic FABP5. Oncotarget 2017, 8, 31041–31056. [Google Scholar] [CrossRef] [Green Version]
- Al-Jameel, W.; Gou, X.; Jin, X.; Zhang, J.; Wei, Q.; Ai, J.; Li, H.; Al-Bayati, A.; Platt-Higgins, A.; Pettitt, A.; et al. Inactivated FABP5 suppresses malignant progression of prostate cancer cells by inhibiting the activation of nuclear fatty acid receptor PPARgamma. Genes Cancer 2019, 10, 80–96. [Google Scholar] [CrossRef] [Green Version]
- Carbonetti, G.; Wilpshaar, T.; Kroonen, J.; Studholme, K.; Converso, C.; d’Oelsnitz, S.; Kaczocha, M. FABP5 coordinates lipid signaling that promotes prostate cancer metastasis. Sci. Rep. 2019, 9, 18944. [Google Scholar] [CrossRef] [Green Version]
- Nitschke, K.; Erben, P.; Waldbillig, F.; Abdelhadi, A.; Weis, C.A.; Gottschalt, M.; Wahby, S.; Nuhn, P.; Boutros, M.; Michel, M.S.; et al. Clinical relevance of gene expression in localized and metastatic prostate cancer exemplified by FABP5. World J. Urol. 2020, 38, 637–645. [Google Scholar] [CrossRef]
- Graf, S.A.; Heppt, M.V.; Wessely, A.; Krebs, S.; Kammerbauer, C.; Hornig, E.; Strieder, A.; Blum, H.; Bosserhoff, A.K.; Berking, C. The myelin protein PMP2 is regulated by SOX10 and drives melanoma cell invasion. Pigm. Cell Melanoma Res. 2019, 32, 424–434. [Google Scholar] [CrossRef]
- Al Fayi, M.S.; Gou, X.; Forootan, S.S.; Al-Jameel, W.; Bao, Z.; Rudland, P.R.; Cornford, P.A.; Hussain, S.A.; Ke, Y. The increased expression of fatty acid-binding protein 9 in prostate cancer and its prognostic significance. Oncotarget 2016, 7, 82783–82797. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Hammamieh, R.; Neill, R.; Melhem, M.; Jett, M. Expression pattern of fatty acid-binding proteins in human normal and cancer prostate cells and tissues. Clin. Cancer Res. 2001, 7, 1706–1715. [Google Scholar] [PubMed]
- Hammamieh, R.; Chakraborty, N.; Das, R.; Jett, M. Molecular impacts of antisense complementary to the liver fatty acid binding protein (FABP) mRNA in DU 145 prostate cancer cells in vitro. J. Exp. Ther. Oncol. 2004, 4, 195–202. [Google Scholar] [PubMed]
- Tolle, A.; Suhail, S.; Jung, M.; Jung, K.; Stephan, C. Fatty acid binding proteins (FABPs) in prostate, bladder and kidney cancer cell lines and the use of IL-FABP as survival predictor in patients with renal cell carcinoma. BMC Cancer 2011, 11, 302. [Google Scholar] [CrossRef] [Green Version]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef] [Green Version]
- Wellen, K.E.; Thompson, C.B. Cellular metabolic stress: Considering how cells respond to nutrient excess. Mol. Cell 2010, 40, 323–332. [Google Scholar] [CrossRef] [Green Version]
- Cruz, A.L.S.; Barreto, E.A.; Fazolini, N.P.B.; Viola, J.P.B.; Bozza, P.T. Lipid droplets: Platforms with multiple functions in cancer hallmarks. Cell Death Dis. 2020, 11, 105. [Google Scholar] [CrossRef] [Green Version]
- Bensaad, K.; Favaro, E.; Lewis, C.A.; Peck, B.; Lord, S.; Collins, J.M.; Pinnick, K.E.; Wigfield, S.; Buffa, F.M.; Li, J.L.; et al. Fatty acid uptake and lipid storage induced by HIF-1alpha contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 2014, 9, 349–365. [Google Scholar] [CrossRef] [Green Version]
- Delikatny, E.J.; Chawla, S.; Leung, D.J.; Poptani, H. MR-visible lipids and the tumor microenvironment. NMR Biomed. 2011, 24, 592–611. [Google Scholar] [CrossRef] [Green Version]
- Menendez, J.A.; Lupu, R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 2007, 7, 763–777. [Google Scholar] [CrossRef]
- Swinnen, J.V.; Brusselmans, K.; Verhoeven, G. Increased lipogenesis in cancer cells: New players, novel targets. Curr. Opin. Clin. Nutr. Metab. Care 2006, 9, 358–365. [Google Scholar] [CrossRef]
- Kuemmerle, N.B.; Rysman, E.; Lombardo, P.S.; Flanagan, A.J.; Lipe, B.C.; Wells, W.A.; Pettus, J.R.; Froehlich, H.M.; Memoli, V.A.; Morganelli, P.M.; et al. Lipoprotein lipase links dietary fat to solid tumor cell proliferation. Mol. Cancer Ther. 2011, 10, 427–436. [Google Scholar] [CrossRef] [Green Version]
- Zaidi, N.; Lupien, L.; Kuemmerle, N.B.; Kinlaw, W.B.; Swinnen, J.V.; Smans, K. Lipogenesis and lipolysis: The pathways exploited by the cancer cells to acquire fatty acids. Prog. Lipid Res. 2013, 52, 585–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gang, X.; Yang, Y.; Zhong, J.; Jiang, K.; Pan, Y.; Karnes, R.J.; Zhang, J.; Xu, W.; Wang, G.; Huang, H. P300 acetyltransferase regulates fatty acid synthase expression, lipid metabolism and prostate cancer growth. Oncotarget 2016, 7, 15135–15149. [Google Scholar] [CrossRef] [PubMed]
- Migita, T.; Ruiz, S.; Fornari, A.; Fiorentino, M.; Priolo, C.; Zadra, G.; Inazuka, F.; Grisanzio, C.; Palescandolo, E.; Shin, E.; et al. Fatty acid synthase: A metabolic enzyme and candidate oncogene in prostate cancer. J. Natl. Cancer Inst. 2009, 101, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Rossi, S.; Graner, E.; Febbo, P.; Weinstein, L.; Bhattacharya, N.; Onody, T.; Bubley, G.; Balk, S.; Loda, M. Fatty acid synthase expression defines distinct molecular signatures in prostate cancer. Mol. Cancer Res. 2003, 1, 707–715. [Google Scholar]
- Chen, M.; Zhang, J.; Sampieri, K.; Clohessy, J.G.; Mendez, L.; Gonzalez-Billalabeitia, E.; Liu, X.S.; Lee, Y.R.; Fung, J.; Katon, J.M.; et al. An aberrant SREBP-dependent lipogenic program promotes metastatic prostate cancer. Nat. Genet. 2018, 50, 206–218. [Google Scholar] [CrossRef] [Green Version]
- Epstein, M.M.; Kasperzyk, J.L.; Mucci, L.A.; Giovannucci, E.; Price, A.; Wolk, A.; Hakansson, N.; Fall, K.; Andersson, S.O.; Andren, O. Dietary fatty acid intake and prostate cancer survival in Orebro County, Sweden. Am. J. Epidemiol. 2012, 176, 240–252. [Google Scholar] [CrossRef] [Green Version]
- Richman, E.L.; Kenfield, S.A.; Chavarro, J.E.; Stampfer, M.J.; Giovannucci, E.L.; Willett, W.C.; Chan, J.M. Fat intake after diagnosis and risk of lethal prostate cancer and all-cause mortality. JAMA Intern. Med. 2013, 173, 1318–1326. [Google Scholar] [CrossRef] [Green Version]
- Allott, E.H.; Masko, E.M.; Freedland, S.J. Obesity and prostate cancer: Weighing the evidence. Eur. Urol. 2013, 63, 800–809. [Google Scholar] [CrossRef] [Green Version]
- Nassar, Z.D.; Aref, A.T.; Miladinovic, D.; Mah, C.Y.; Raj, G.V.; Hoy, A.J.; Butler, L.M. Peri-prostatic adipose tissue: The metabolic microenvironment of prostate cancer. BJU Int. 2018, 121 (Suppl. 3), 9–21. [Google Scholar] [CrossRef] [Green Version]
- Watt, M.J.; Clark, A.K.; Selth, L.A.; Haynes, V.R.; Lister, N.; Rebello, R.; Porter, L.H.; Niranjan, B.; Whitby, S.T.; Lo, J.; et al. Suppressing fatty acid uptake has therapeutic effects in preclinical models of prostate cancer. Sci. Transl. Med. 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Schaffler, A.; Scholmerich, J.; Buechler, C. Mechanisms of disease: Adipokines and breast cancer - endocrine and paracrine mechanisms that connect adiposity and breast cancer. Nat. Clin. Pract. Endocrinol. Metab. 2007, 3, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Chiang, C.Y.; Daifotis, H.A.; Nieman, K.M.; Fahrmann, J.F.; Lastra, R.R.; Romero, I.L.; Fiehn, O.; Lengyel, E. Adipocyte-Induced FABP4 Expression in Ovarian Cancer Cells Promotes Metastasis and Mediates Carboplatin Resistance. Cancer Res. 2020, 80, 1748–1761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, M.; Narita, S.; Inoue, T.; Koizumi, A.; Saito, M.; Tsuruta, H.; Numakura, K.; Satoh, S.; Nanjo, H.; Sasaki, T.; et al. Fatty acid binding protein 4 enhances prostate cancer progression by upregulating matrix metalloproteinases and stromal cell cytokine production. Oncotarget 2017, 8, 111780–111794. [Google Scholar] [CrossRef] [Green Version]
- Senga, S.; Kobayashi, N.; Kawaguchi, K.; Ando, A.; Fujii, H. Fatty acid-binding protein 5 (FABP5) promotes lipolysis of lipid droplets, de novo fatty acid (FA) synthesis and activation of nuclear factor-kappa B (NF-kappaB) signaling in cancer cells. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 1057–1067. [Google Scholar] [CrossRef]
- Powell, C.A.; Nasser, M.W.; Zhao, H.; Wochna, J.C.; Zhang, X.; Shapiro, C.; Shilo, K.; Ganju, R.K. Fatty acid binding protein 5 promotes metastatic potential of triple negative breast cancer cells through enhancing epidermal growth factor receptor stability. Oncotarget 2015, 6, 6373–6385. [Google Scholar] [CrossRef] [Green Version]
- Ackerman, D.; Simon, M.C. Hypoxia, lipids, and cancer: Surviving the harsh tumor microenvironment. Trends Cell Biol. 2014, 24, 472–478. [Google Scholar] [CrossRef] [Green Version]
- Beloribi-Djefaflia, S.; Vasseur, S.; Guillaumond, F. Lipid metabolic reprogramming in cancer cells. Oncogenesis 2016, 5, e189. [Google Scholar] [CrossRef]
- Biswas, S.; Lunec, J.; Bartlett, K. Non-glucose metabolism in cancer cells--is it all in the fat? Cancer Metast. Rev. 2012, 31, 689–698. [Google Scholar] [CrossRef]
- Luo, X.; Cheng, C.; Tan, Z.; Li, N.; Tang, M.; Yang, L.; Cao, Y. Emerging roles of lipid metabolism in cancer metastasis. Mol. Cancer 2017, 16, 76. [Google Scholar] [CrossRef] [Green Version]
- Buschemeyer, W.C., 3rd; Freedland, S.J. Obesity and prostate cancer: Epidemiology and clinical implications. Eur. Urol. 2007, 52, 331–343. [Google Scholar] [CrossRef]
- Chavarro, J.E.; Stampfer, M.J.; Li, H.; Campos, H.; Kurth, T.; Ma, J. A prospective study of polyunsaturated fatty acid levels in blood and prostate cancer risk. Cancer Epidemiol. Biom. Prev. 2007, 16, 1364–1370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Daniels, G.; Lee, P.; Monaco, M.E. Lipid metabolism in prostate cancer. Am. J. Clin. Exp. Urol. 2014, 2, 111–120. [Google Scholar] [PubMed]
- Pascual, G.; Dominguez, D.; Benitah, S.A. The contributions of cancer cell metabolism to metastasis. Dis. Model Mech. 2018, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Kang, Y. Lipid Metabolism Fuels Cancer’s Spread. Cell Metab. 2017, 25, 228–230. [Google Scholar] [CrossRef] [Green Version]
- Tousignant, K.D.; Rockstroh, A.; Taherian Fard, A.; Lehman, M.L.; Wang, C.; McPherson, S.J.; Philp, L.K.; Bartonicek, N.; Dinger, M.E.; Nelson, C.C.; et al. Lipid Uptake Is an Androgen-Enhanced Lipid Supply Pathway Associated with Prostate Cancer Disease Progression and Bone Metastasis. Mol. Cancer Res. 2019, 17, 1166–1179. [Google Scholar] [CrossRef] [Green Version]
- Clarke, N.W.; Brown, M.D. The influence of lipid metabolism on prostate cancer development and progression: Is it time for a closer look? Eur. Urol. 2007, 52, 3–4. [Google Scholar] [CrossRef]
- Gu, Z.; Suburu, J.; Chen, H.; Chen, Y.Q. Mechanisms of omega-3 polyunsaturated fatty acids in prostate cancer prevention. Biomed. Res. Int. 2013, 2013, 824563. [Google Scholar] [CrossRef] [Green Version]
- Li, C.C.; Hou, Y.C.; Yeh, C.L.; Yeh, S.L. Effects of eicosapentaenoic acid and docosahexaenoic acid on prostate cancer cell migration and invasion induced by tumor-associated macrophages. PLoS ONE 2014, 9, e99630. [Google Scholar] [CrossRef] [Green Version]
- Sikka, S.; Chen, L.; Sethi, G.; Kumar, A.P. Targeting PPARgamma Signaling Cascade for the Prevention and Treatment of Prostate Cancer. PPAR Res. 2012, 2012, 968040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elix, C.; Pal, S.K.; Jones, J.O. The role of peroxisome proliferator-activated receptor gamma in prostate cancer. Asian J. Androl. 2018, 20, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Senga, S.; Kawaguchi, K.; Kobayashi, N.; Ando, A.; Fujii, H. A novel fatty acid-binding protein 5-estrogen-related receptor alpha signaling pathway promotes cell growth and energy metabolism in prostate cancer cells. Oncotarget 2018, 9, 31753–31770. [Google Scholar] [CrossRef] [PubMed]
- Furuhashi, M. Fatty Acid-Binding Protein 4 in Cardiovascular and Metabolic Diseases. J. Atheroscl. Thromb. 2019, 26, 216–232. [Google Scholar] [CrossRef] [Green Version]
- Nieman, K.M.; Kenny, H.A.; Penicka, C.V.; Ladanyi, A.; Buell-Gutbrod, R.; Zillhardt, M.R.; Romero, I.L.; Carey, M.S.; Mills, G.B.; Hotamisligil, G.S.; et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 2011, 17, 1498–1503. [Google Scholar] [CrossRef] [Green Version]
- Herroon, M.K.; Rajagurubandara, E.; Hardaway, A.L.; Powell, K.; Turchick, A.; Feldmann, D.; Podgorski, I. Bone marrow adipocytes promote tumor growth in bone via FABP4-dependent mechanisms. Oncotarget 2013, 4, 2108–2123. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, I.; Mui, E.; Galbraith, L.; Patel, R.; Tan, E.H.; Salji, M.; Rust, A.G.; Repiscak, P.; Hedley, A.; Markert, E.; et al. Sleeping Beauty screen reveals Pparg activation in metastatic prostate cancer. Proc. Natl. Acad. Sci. USA 2016, 113, 8290–8295. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Tirinato, L.; Pagliari, F.; Limongi, T.; Marini, M.; Falqui, A.; Seco, J.; Candeloro, P.; Liberale, C.; Di Fabrizio, E. An Overview of Lipid Droplets in Cancer and Cancer Stem Cells. Stem Cells Int. 2017, 2017, 1656053. [Google Scholar] [CrossRef]
- Morandi, A.; Taddei, M.L.; Chiarugi, P.; Giannoni, E. Targeting the Metabolic Reprogramming That Controls Epithelial-to-Mesenchymal Transition in Aggressive Tumors. Front. Oncol. 2017, 7, 40. [Google Scholar] [CrossRef] [Green Version]
- Bolos, V.; Peinado, H.; Perez-Moreno, M.A.; Fraga, M.F.; Esteller, M.; Cano, A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: A comparison with Snail and E47 repressors. J. Cell Sci. 2003, 116, 499–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, S.H.M.; Fang, C.M.; Chuah, L.H.; Leong, C.O.; Ngai, S.C. E-cadherin: Its dysregulation in carcinogenesis and clinical implications. Crit. Rev. Oncol. Hematol. 2018, 121, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Liu, Z.; Niu, B.; Zhang, J.; Tan, T.K.; Lee, S.R.; Zhao, Y.; Harris, D.C.; Zheng, G. E-cadherin/beta-catenin complex and the epithelial barrier. J. Biomed. Biotechnol. 2011, 2011, 567305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levi, L.; Lobo, G.; Doud, M.K.; von Lintig, J.; Seachrist, D.; Tochtrop, G.P.; Noy, N. Genetic ablation of the fatty acid-binding protein FABP5 suppresses HER2-induced mammary tumorigenesis. Cancer Res. 2013, 73, 4770–4780. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; Li, Y.; Xing, L.; Tan, Y.; Sun, J.; Zeng, B.; Xiang, T.; Tan, J.; Ren, G.; Wang, Y. Utilization of adipocyte-derived lipids and enhanced intracellular trafficking of fatty acids contribute to breast cancer progression. Cell Commun. Signal 2018, 16, 32. [Google Scholar] [CrossRef] [Green Version]
- Ohata, T.; Yokoo, H.; Kamiyama, T.; Fukai, M.; Aiyama, T.; Hatanaka, Y.; Hatanaka, K.; Wakayama, K.; Orimo, T.; Kakisaka, T.; et al. Fatty acid-binding protein 5 function in hepatocellular carcinoma through induction of epithelial-mesenchymal transition. Cancer Med. 2017, 6, 1049–1061. [Google Scholar] [CrossRef]
- Jin, J.; Zhang, Z.; Zhang, S.; Chen, X.; Chen, Z.; Hu, P.; Wang, J.; Xie, C. Fatty acid binding protein 4 promotes epithelial-mesenchymal transition in cervical squamous cell carcinoma through AKT/GSK3beta/Snail signaling pathway. Mol. Cell Endocrinol. 2018, 461, 155–164. [Google Scholar] [CrossRef]
- Takahashi, H.; McCaffery, J.M.; Irizarry, R.A.; Boeke, J.D. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol. Cell 2006, 23, 207–217. [Google Scholar] [CrossRef]
- Wellen, K.E.; Hatzivassiliou, G.; Sachdeva, U.M.; Bui, T.V.; Cross, J.R.; Thompson, C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 2009, 324, 1076–1080. [Google Scholar] [CrossRef] [Green Version]
- McDonnell, E.; Crown, S.B.; Fox, D.B.; Kitir, B.; Ilkayeva, O.R.; Olsen, C.A.; Grimsrud, P.A.; Hirschey, M.D. Lipids Reprogram Metabolism to Become a Major Carbon Source for Histone Acetylation. Cell Rep. 2016, 17, 1463–1472. [Google Scholar] [CrossRef] [Green Version]
- Kong, D.; Ahmad, A.; Bao, B.; Li, Y.; Banerjee, S.; Sarkar, F.H. Histone deacetylase inhibitors induce epithelial-to-mesenchymal transition in prostate cancer cells. PLoS ONE 2012, 7, e45045. [Google Scholar] [CrossRef] [PubMed]
- Mobley, R.J.; Abell, A.N. Controlling Epithelial to Mesenchymal Transition through Acetylation of Histone H2BK5. J. Nat. Sci. 2017, 3, e432. [Google Scholar] [PubMed]
- Zadra, G.; Ribeiro, C.F.; Chetta, P.; Ho, Y.; Cacciatore, S.; Gao, X.; Syamala, S.; Bango, C.; Photopoulos, C.; Huang, Y.; et al. Inhibition of de novo lipogenesis targets androgen receptor signaling in castration-resistant prostate cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 631–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bielenberg, D.R.; Zetter, B.R. The Contribution of Angiogenesis to the Process of Metastasis. Cancer J. 2015, 21, 267–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Treps, L.; Conradi, L.C.; Harjes, U.; Carmeliet, P. Manipulating Angiogenesis by Targeting Endothelial Metabolism: Hitting the Engine Rather than the Drivers-A New Perspective? Pharmacol. Rev. 2016, 68, 872–887. [Google Scholar] [CrossRef] [Green Version]
- Melegh, Z.; Oltean, S. Targeting Angiogenesis in Prostate Cancer. Int. J. Mol. Sci. 2019, 20, 2676. [Google Scholar] [CrossRef] [Green Version]
- Baenke, F.; Peck, B.; Miess, H.; Schulze, A. Hooked on fat: The role of lipid synthesis in cancer metabolism and tumour development. Dis. Model Mech. 2013, 6, 1353–1363. [Google Scholar] [CrossRef] [Green Version]
- Iwamoto, H.; Abe, M.; Yang, Y.; Cui, D.; Seki, T.; Nakamura, M.; Hosaka, K.; Lim, S.; Wu, J.; He, X.; et al. Cancer Lipid Metabolism Confers Antiangiogenic Drug Resistance. Cell Metab. 2018, 28, 104–117. [Google Scholar] [CrossRef] [Green Version]
- Masouye, I.; Hagens, G.; Van Kuppevelt, T.H.; Madsen, P.; Saurat, J.H.; Veerkamp, J.H.; Pepper, M.S.; Siegenthaler, G. Endothelial cells of the human microvasculature express epidermal fatty acid-binding protein. Circ. Res. 1997, 81, 297–303. [Google Scholar] [CrossRef] [Green Version]
- Jing, C.; Beesley, C.; Foster, C.S.; Chen, H.; Rudland, P.S.; West, D.C.; Fujii, H.; Smith, P.H.; Ke, Y. Human cutaneous fatty acid-binding protein induces metastasis by up-regulating the expression of vascular endothelial growth factor gene in rat Rama 37 model cells. Cancer Res. 2001, 61, 4357–4364. [Google Scholar]
- Jing, C.; Beesley, C.; Foster, C.S.; Rudland, P.S.; Fujii, H.; Ono, T.; Chen, H.; Smith, P.H.; Ke, Y. Identification of the messenger RNA for human cutaneous fatty acid-binding protein as a metastasis inducer. Cancer Res. 2000, 60, 2390–2398. [Google Scholar]
- Cataltepe, O.; Arikan, M.C.; Ghelfi, E.; Karaaslan, C.; Ozsurekci, Y.; Dresser, K.; Li, Y.; Smith, T.W.; Cataltepe, S. Fatty acid binding protein 4 is expressed in distinct endothelial and non-endothelial cell populations in glioblastoma. Neuropathol. Appl. Neurobiol. 2012, 38, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Cataltepe, S.; Arikan, M.C.; Liang, X.; Smith, T.W.; Cataltepe, O. Fatty acid binding protein 4 expression in cerebral vascular malformations: Implications for vascular remodelling. Neuropathol. Appl. Neurobiol. 2015, 41, 646–656. [Google Scholar] [CrossRef] [PubMed]
- Elmasri, H.; Ghelfi, E.; Yu, C.W.; Traphagen, S.; Cernadas, M.; Cao, H.; Shi, G.P.; Plutzky, J.; Sahin, M.; Hotamisligil, G.; et al. Endothelial cell-fatty acid binding protein 4 promotes angiogenesis: Role of stem cell factor/c-kit pathway. Angiogenesis 2012, 15, 457–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elmasri, H.; Karaaslan, C.; Teper, Y.; Ghelfi, E.; Weng, M.; Ince, T.A.; Kozakewich, H.; Bischoff, J.; Cataltepe, S. Fatty acid binding protein 4 is a target of VEGF and a regulator of cell proliferation in endothelial cells. Faseb J. 2009, 23, 3865–3873. [Google Scholar] [CrossRef] [Green Version]
- Ghelfi, E.; Yu, C.W.; Elmasri, H.; Terwelp, M.; Lee, C.G.; Bhandari, V.; Comhair, S.A.; Erzurum, S.C.; Hotamisligil, G.S.; Elias, J.A.; et al. Fatty acid binding protein 4 regulates VEGF-induced airway angiogenesis and inflammation in a transgenic mouse model: Implications for asthma. Am. J. Pathol. 2013, 182, 1425–1433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, C.W.; Liang, X.; Lipsky, S.; Karaaslan, C.; Kozakewich, H.; Hotamisligil, G.S.; Bischoff, J.; Cataltepe, S. Dual role of fatty acid-binding protein 5 on endothelial cell fate: A potential link between lipid metabolism and angiogenic responses. Angiogenesis 2016, 19, 95–106. [Google Scholar] [CrossRef] [Green Version]
- Pan, L.; Xiao, H.; Liao, R.; Chen, Q.; Peng, C.; Zhang, Y.; Mu, T.; Wu, Z. Fatty acid binding protein 5 promotes tumor angiogenesis and activates the IL6/STAT3/VEGFA pathway in hepatocellular carcinoma. Biomed. Pharmacother. 2018, 106, 68–76. [Google Scholar] [CrossRef]
- Levi, L.; Wang, Z.; Doud, M.K.; Hazen, S.L.; Noy, N. Saturated fatty acids regulate retinoic acid signalling and suppress tumorigenesis by targeting fatty acid-binding protein 5. Nat. Commun. 2015, 6, 8794. [Google Scholar] [CrossRef] [Green Version]
- Harjes, U.; Bridges, E.; Gharpure, K.M.; Roxanis, I.; Sheldon, H.; Miranda, F.; Mangala, L.S.; Pradeep, S.; Lopez-Berestein, G.; Ahmed, A.; et al. Antiangiogenic and tumour inhibitory effects of downregulating tumour endothelial FABP4. Oncogene 2017, 36, 912–921. [Google Scholar] [CrossRef] [Green Version]
- Tan, N.S.; Shaw, N.S.; Vinckenbosch, N.; Liu, P.; Yasmin, R.; Desvergne, B.; Wahli, W.; Noy, N. Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol. Cell. Biol. 2002, 22, 5114–5127. [Google Scholar] [CrossRef] [Green Version]
- Kannan-Thulasiraman, P.; Seachrist, D.D.; Mahabeleshwar, G.H.; Jain, M.K.; Noy, N. Fatty acid-binding protein 5 and PPARbeta/delta are critical mediators of epidermal growth factor receptor-induced carcinoma cell growth. J. Biol. Chem. 2010, 285, 19106–19115. [Google Scholar] [CrossRef] [Green Version]
- Boss, M.; Kemmerer, M.; Brune, B.; Namgaladze, D. FABP4 inhibition suppresses PPARgamma activity and VLDL-induced foam cell formation in IL-4-polarized human macrophages. Atherosclerosis 2015, 240, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Ahmadian, M.; Suh, J.M.; Hah, N.; Liddle, C.; Atkins, A.R.; Downes, M.; Evans, R.M. PPARgamma signaling and metabolism: The good, the bad and the future. Nat. Med. 2013, 19, 557–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Colby, J.K.; Zuo, X.; Jaoude, J.; Wei, D.; Shureiqi, I. The Role of PPAR-delta in Metabolism, Inflammation, and Cancer: Many Characters of a Critical Transcription Factor. Int. J. Mol. Sci. 2018, 19, 1339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dupuy, F.; Tabaries, S.; Andrzejewski, S.; Dong, Z.; Blagih, J.; Annis, M.G.; Omeroglu, A.; Gao, D.; Leung, S.; Amir, E.; et al. PDK1-Dependent Metabolic Reprogramming Dictates Metastatic Potential in Breast Cancer. Cell Metab. 2015, 22, 577–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brasaemle, D.L.; Barber, T.; Wolins, N.E.; Serrero, G.; Blanchette-Mackie, E.J.; Londos, C. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J. Lipid Res. 1997, 38, 2249–2263. [Google Scholar]
- Carmeliet, P. VEGF as a key mediator of angiogenesis in cancer. Oncology (Williston Park) 2005, 69 (Suppl. 3), 4–10. [Google Scholar] [CrossRef]
- Forootan, F.S.; Forootan, S.S.; Gou, X.; Yang, J.; Liu, B.; Chen, D.; Al Fayi, M.S.; Al-Jameel, W.; Rudland, P.S.; Hussain, S.A.; et al. Fatty acid activated PPARgamma promotes tumorigenicity of prostate cancer cells by up regulating VEGF via PPAR responsive elements of the promoter. Oncotarget 2016, 7, 9322–9339. [Google Scholar] [CrossRef] [Green Version]
- Ha, X.; Wang, J.; Chen, K.; Deng, Y.; Zhang, X.; Feng, J.; Li, X.; Zhu, J.; Ma, Y.; Qiu, T.; et al. Free Fatty Acids Promote the Development of Prostate Cancer by Upregulating Peroxisome Proliferator-Activated Receptor Gamma. Cancer Manag. Res. 2020, 12, 1355–1369. [Google Scholar] [CrossRef] [Green Version]
- Nomura, D.K.; Long, J.Z.; Niessen, S.; Hoover, H.S.; Ng, S.W.; Cravatt, B.F. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell 2010, 140, 49–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tontonoz, P.; Hu, E.; Spiegelman, B.M. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 1994, 79, 1147–1156. [Google Scholar] [CrossRef]
- Adida, A.; Spener, F. Adipocyte-type fatty acid-binding protein as inter-compartmental shuttle for peroxisome proliferator activated receptor gamma agonists in cultured cell. Biochim. Biophys. Acta 2006, 1761, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Carbonetti, G.; Converso, C.; Clement, T.; Wang, C.; Trotman, L.C.; Ojima, I.; Kaczocha, M. Docetaxel/cabazitaxel and fatty acid binding protein 5 inhibitors produce synergistic inhibition of prostate cancer growth. Prostate 2020, 80, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Harraz, A.M.; Atia, N.; Ismail, A.; Shady, A.; Farg, H.; Gabr, H.; Fouda, M.; Abol-Enein, H.; Abdel-Aziz, A.F. Evaluation of serum fatty acid binding protein-4 (FABP-4) as a novel biomarker to predict biopsy outcomes in prostate biopsy naive patients. Int. Urol. Nephrol. 2020, 52, 1483–1490. [Google Scholar] [CrossRef]

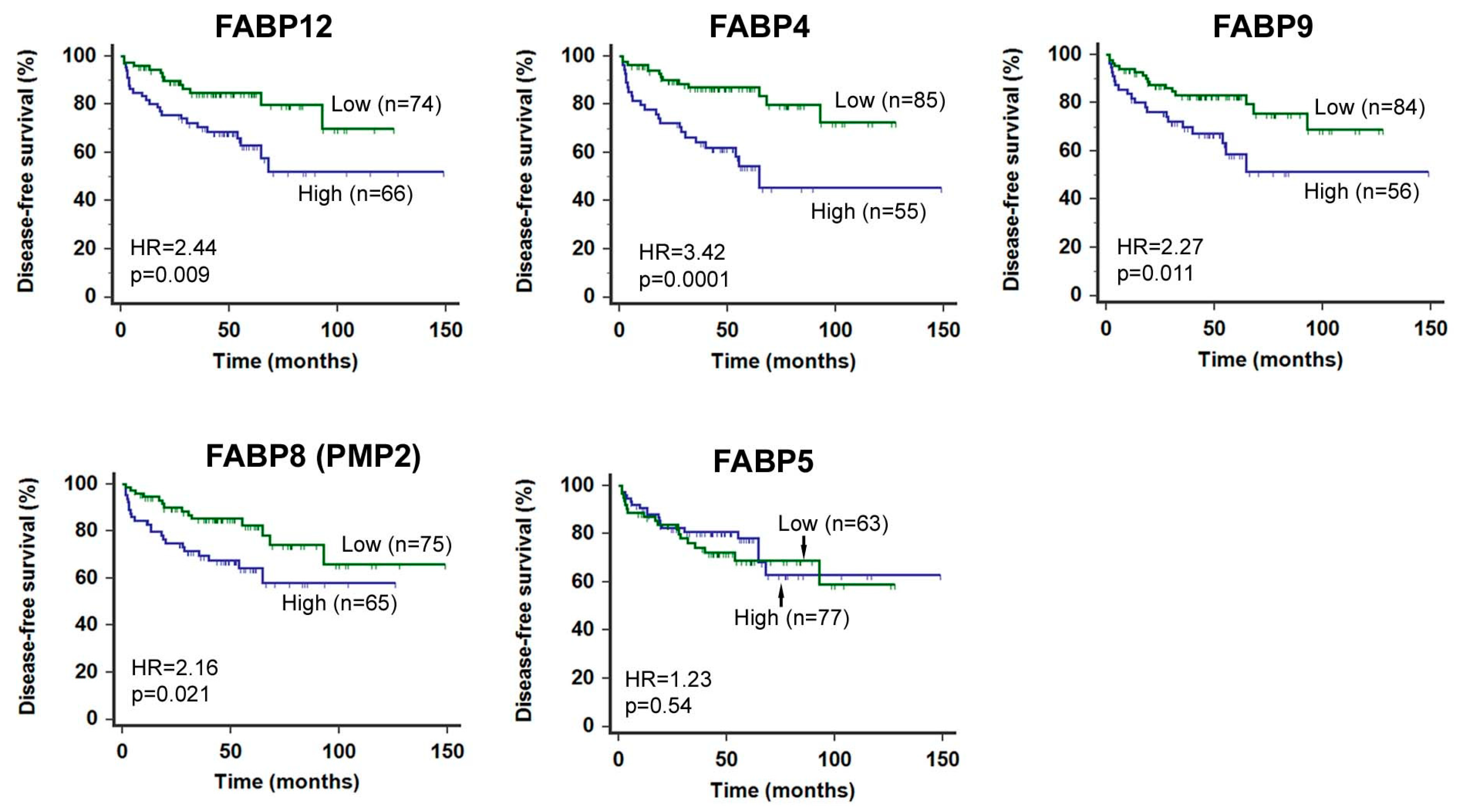
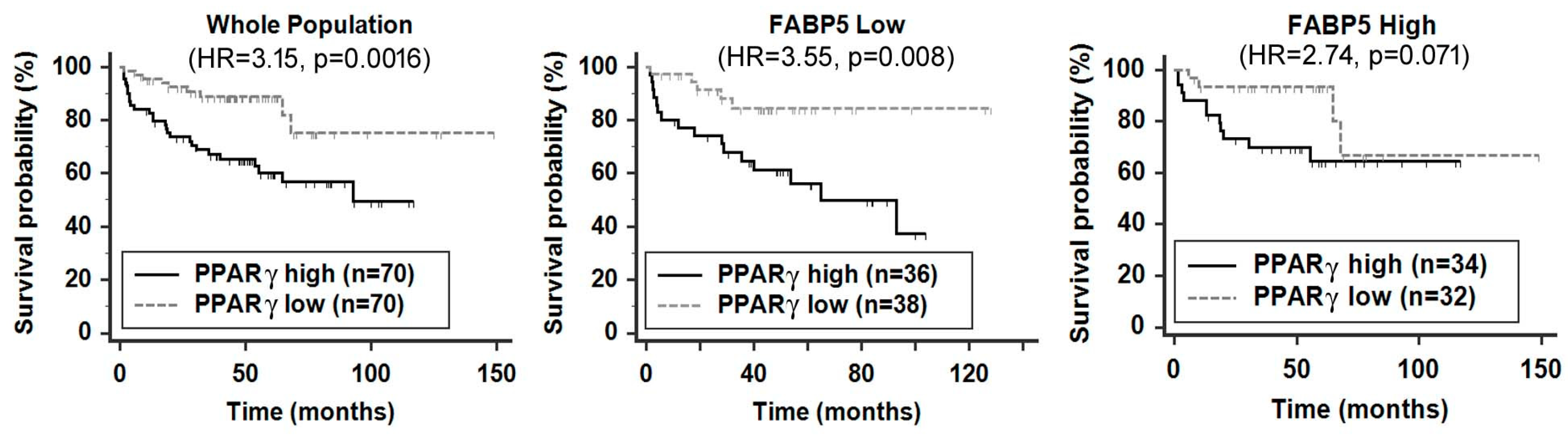

| Outcome | Tumor Class | FABP12 | FABP4 | FABP9 | FABP8 | FABP5 |
|---|---|---|---|---|---|---|
| Recurrence | Recurrence-free (n = 104) | −0.012 ± 0.93 | −0.337 ± 1.02 | −0.144 ± 0.83 | −0.214 ± 1.05 | 2.290 ± 3.90 |
| Recurred (n = 36) | 0.237 ± 1.02 | 0.461 ± 1.45 | 0.261 ± 1.29 | 0.220 ± 1.51 | 2.148 ± 3.63 | |
| p value | 0.178 | <0.001 | 0.033 | 0.061 | 0.849 | |
| Gleason score | Grade 6 (n = 41) | 0.109 ± 0.99 | −0.224 ± 1.01 | −0.010 ± 0.88 | −0.164 ± 1.12 | 2.706 ± 3.99 |
| Grade 7 (n = 76) | −0.123 ± 0.84 | −0.309 ± 1.07 | −0.262 ± 0.80 | −0.269 ± 0.99 | 2.090 ± 3.94 | |
| Grade 8 (n = 11) | 0.220 ± 1.05 | −0.240 ± 0.933 | 0.157 ± 0.89 | −0.041 ± 1.43 | 1.819 ± 3.57 | |
| Grade 9 (n = 11) | 0.910 ± 1.15 | 1.459 ± 1.43 | 1.000 ± 1.29 | 0.998 ± 1.53 | 2.785 ± 2.62 | |
| p value | 0.007 | <0.001 | <0.001 | 0.007 | 0.793 |
| Genes | FABP12 | FABP4 | FABP9 | FABP8 |
|---|---|---|---|---|
| FABP4 | 0.62 (p < 0.0001) | |||
| FABP9 | 0.71 (p < 0.0001) | 0.64 (p < 0.0001) | ||
| FABP8 | 0.55 (p < 0.0001) | 0.55 (p < 0.0001) | 0.69 (p < 0.0001) | |
| FABP5 | 0.15 (p = 0.07) | 0.02 (p = 0.85) | 0.17 (p = 0.03) | 0.06 (p = 0.46) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, R.-Z.; Godbout, R. An Amplified Fatty Acid-Binding Protein Gene Cluster in Prostate Cancer: Emerging Roles in Lipid Metabolism and Metastasis. Cancers 2020, 12, 3823. https://doi.org/10.3390/cancers12123823
Liu R-Z, Godbout R. An Amplified Fatty Acid-Binding Protein Gene Cluster in Prostate Cancer: Emerging Roles in Lipid Metabolism and Metastasis. Cancers. 2020; 12(12):3823. https://doi.org/10.3390/cancers12123823
Chicago/Turabian StyleLiu, Rong-Zong, and Roseline Godbout. 2020. "An Amplified Fatty Acid-Binding Protein Gene Cluster in Prostate Cancer: Emerging Roles in Lipid Metabolism and Metastasis" Cancers 12, no. 12: 3823. https://doi.org/10.3390/cancers12123823







