Novel Semi-Replicative Retroviral Vector Mediated Double Suicide Gene Transfer Enhances Antitumor Effects in Patient-Derived Glioblastoma Models
Abstract
1. Introduction
2. Results
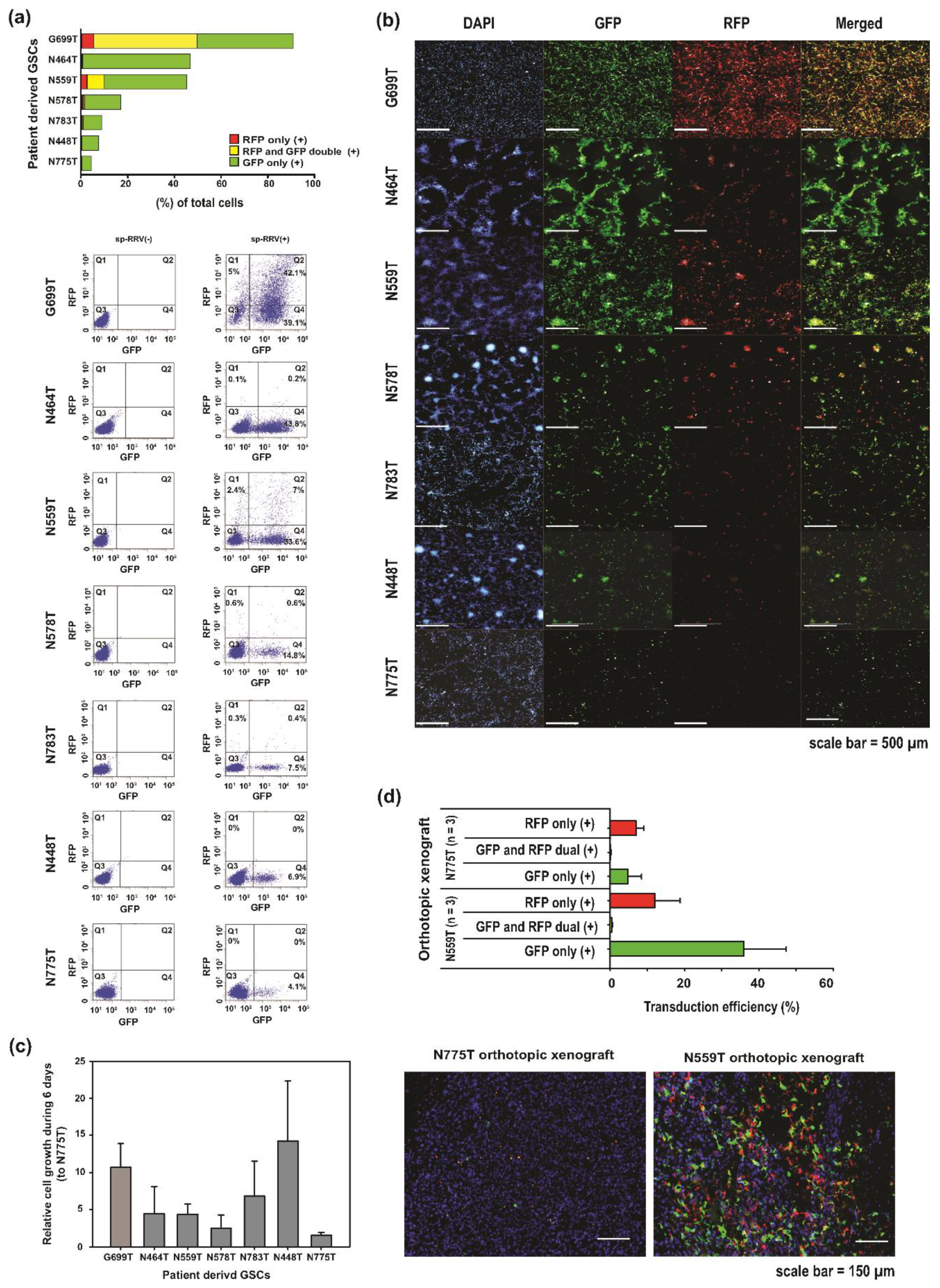
2.1. Analysis of sp-RRV Dissemination In Vitro in Patient-Derived GSCs
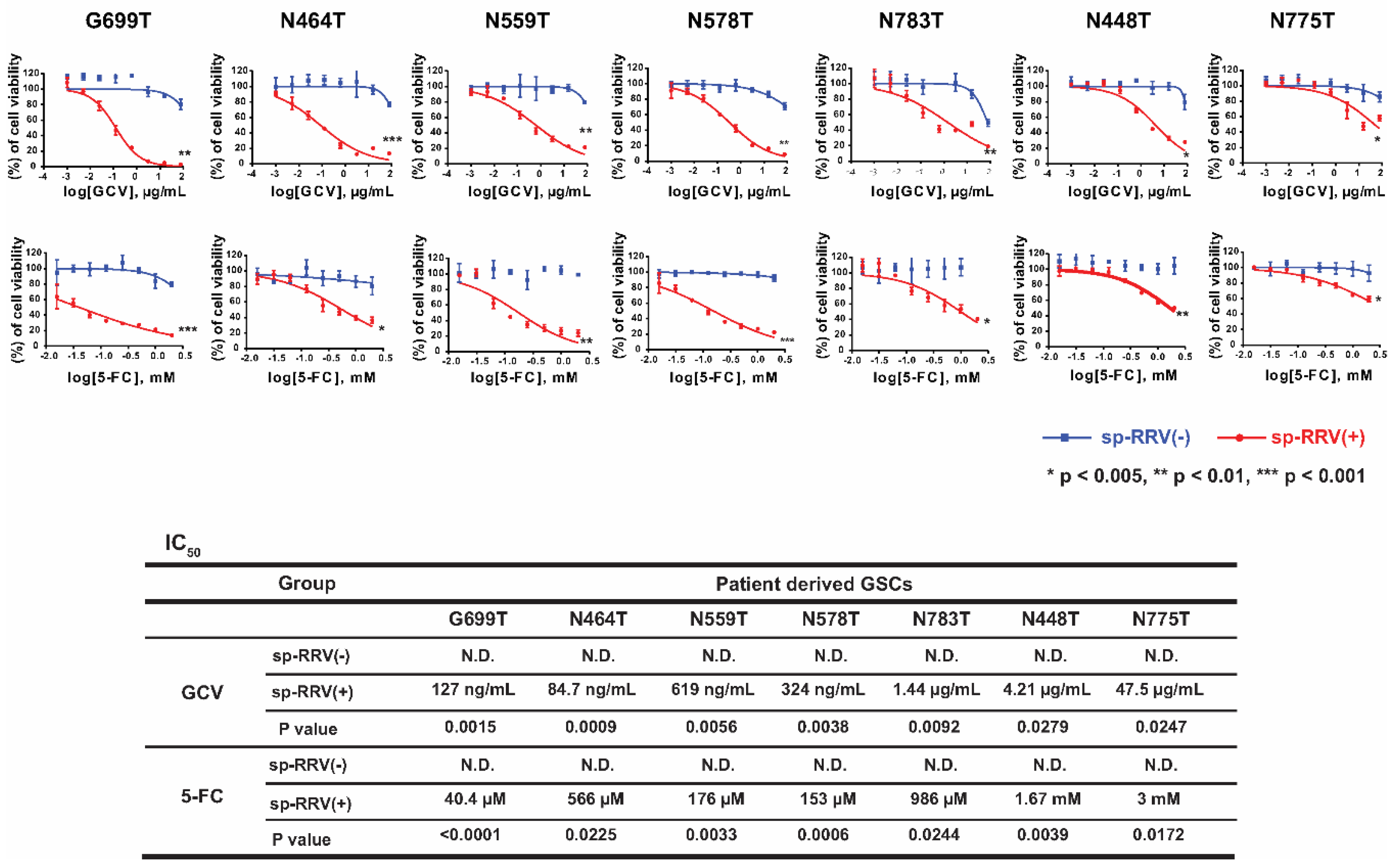
2.2. Evaluation of 5-FC and GCV Sensitivity in Infected and Bystander Cells after TK and CD Delivery In Vitro
2.3. Synergistic Anti-Tumor Effects of sp-RRVe-eEF1α-TK and sRRVgp-eEF1α-CD after Treatment with GCV and 5-FC in Glioblastoma Patient-Derived Orthotopic Xenografts
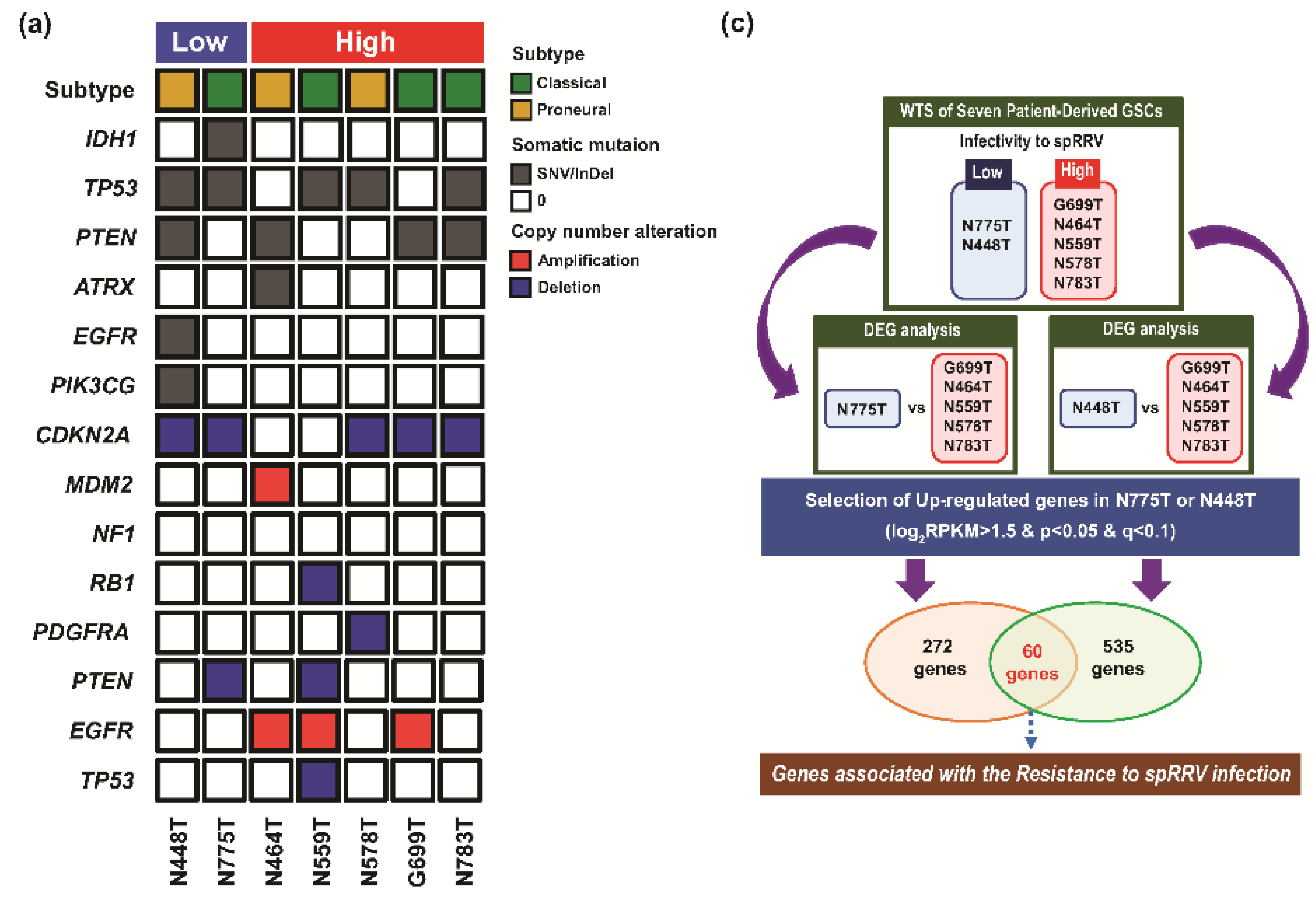
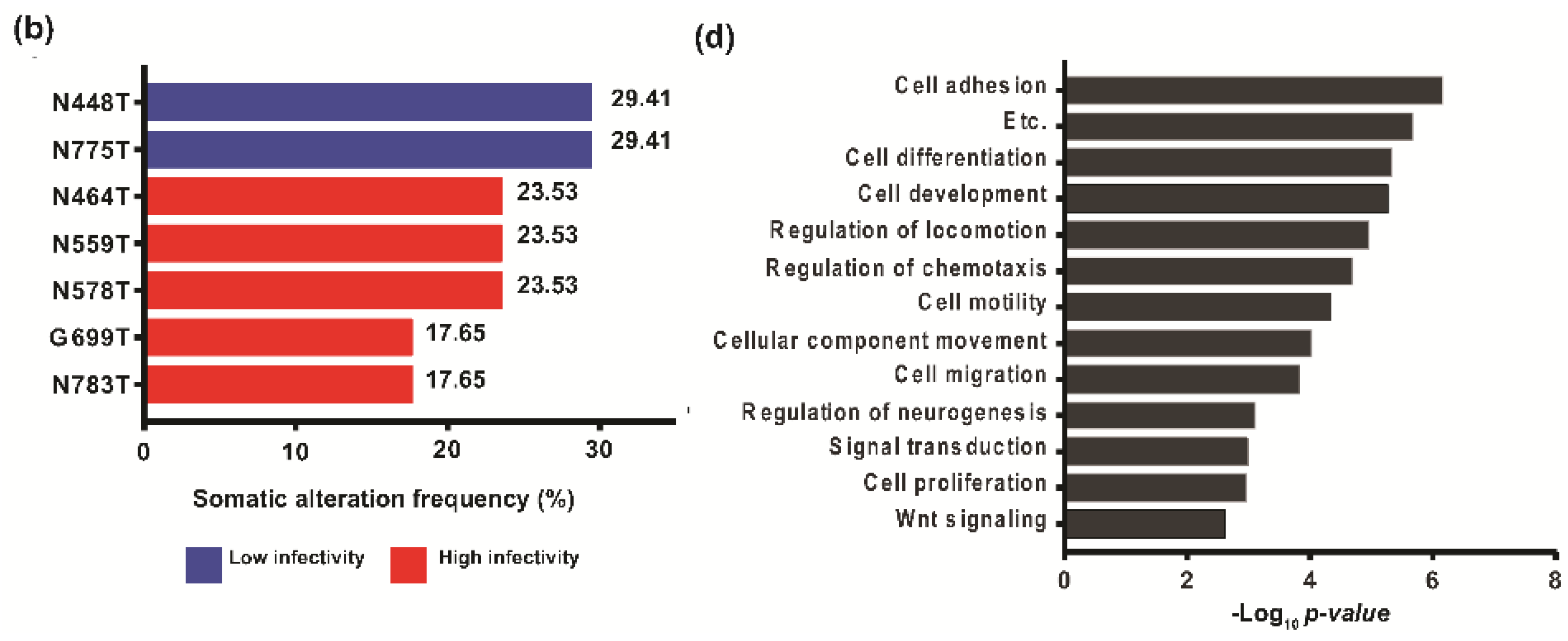
2.4. Determination of Host-Specific Inhibitory Factors to the sp-RRV System through Comprehensive Genomic Profiling of Patient-Derived GSCs
3. Discussion
4. Materials and Methods
4.1. Patient-Derived GSC Establishment
4.2. Sphere-Forming Culture of Patient-Derived GSCs
4.3. Drugs and Reagents
4.4. sp-RRV Production
4.5. sp-RRV System Transduction of GSCs
4.6. Determination of the Transduction Efficiency of the Patient-Derived GSC sp-RRV System Using FACS Analysis and Operetta High-Content Imaging System In Vitro
4.7. In Vivo Transduction Efficacy of Orthotopic Xenografts Based on Patient-Derived GSCs and sp-RRV System
4.8. Cell Proliferation Assay
4.9. In Vitro GCV and 5-FC Sensitivity Assay of sp-RRV-Transduced Patient-Derived GSCs
4.10. Validation of sp-RRV Therapeutic Efficacy in Orthotopic Xenografts
4.11. Histological and IHC Analysis
4.12. Quantitative Analysis of IHC Staining
4.13. Targeted-Panel Sequencing via GliomaSCAN™
4.14. WTS
4.15. DEG and GO Analysis of DEGs
4.16. Statistical Analysis
4.17. Data Access
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Facchino, S.; Abdouh, M.; Bernier, G. Brain cancer stem cells: Current status on glioblastoma multiforme. Cancers 2011, 3, 1777–1797. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Tso, J.; Ye, F.; Choe, J.; Liu, Y.; Liau, L.M.; Tso, C.L. Essential gene pathways for glioblastoma stem cells: Clinical implications for prevention of tumor recurrence. Cancers 2011, 3, 1975–1995. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Lee, K.; Kim, D.G.; Yang, H.; Nam, D.H. Facilitating tailored therapeutic strategies for glioblastoma through an orthotopic patient-derived xenograft platform. Histol. Histopathol. 2016, 31, 269–283. [Google Scholar] [PubMed]
- Patrizii, M.; Bartucci, M.; Pine, S.R.; Sabaawy, H.E. Utility of Glioblastoma Patient-Derived Orthotopic Xenografts in Drug Discovery and Personalized Therapy. Front. Oncol. 2018, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Gillet, J.P.; Varma, S.; Gottesman, M.M. The clinical relevance of cancer cell lines. J. Natl. Cancer Inst. 2013, 105, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Wang, J.; Sa, J.K.; Ladewig, E.; Lee, H.O.; Lee, I.H.; Kang, H.J.; Rosenbloom, D.S.; Camara, P.G.; Liu, Z.; et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat. Genet. 2017, 49, 594–599. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Liu, Z.; Sa, J.K.; Shin, S.; Wang, J.; Bordyuh, M.; Cho, H.J.; Elliott, O.; Chu, T.; Choi, S.W.; et al. Pharmacogenomic landscape of patient-derived tumor cells informs precision oncology therapy. Nat. Genet. 2018, 50, 1399–1411. [Google Scholar] [CrossRef] [PubMed]
- Ginn, S.L.; Amaya, A.K.; Alexander, I.E.; Edelstein, M.; Abedi, M.R. Gene therapy clinical trials worldwide to 2017: An update. J. Gene. Med. 2018, 20, e3015. [Google Scholar] [CrossRef]
- Dixit, K.; Kumthekar, P. Gene Delivery in Neuro-Oncology. Curr. Oncol. Rep. 2017, 19, 69. [Google Scholar] [CrossRef]
- Kubo, S.; Takagi-Kimura, M.; Logg, C.R.; Kasahara, N. Highly efficient tumor transduction and antitumor efficacy in experimental human malignant mesothelioma using replicating gibbon ape leukemia virus. Cancer Gene. Ther. 2013, 20, 671–677. [Google Scholar] [CrossRef][Green Version]
- Logg, C.R.; Baranick, B.T.; Lemp, N.A.; Kasahara, N. Adaptive evolution of a tagged chimeric gammaretrovirus: Identification of novel cis-acting elements that modulate splicing. J. Mol. Biol. 2007, 369, 1214–1229. [Google Scholar] [CrossRef]
- Miller, D.G.; Adam, M.A.; Miller, A.D. Gene transfer by retrovirus vectors occurs only in cells that are actively replicating at the time of infection. Mol. Cell Biol. 1990, 10, 4239–4242. [Google Scholar] [CrossRef]
- Dalba, C.; Bellier, B.; Kasahara, N.; Klatzmann, D. Replication-competent vectors and empty virus-like particles: New retroviral vector designs for cancer gene therapy or vaccines. Mol. Ther. 2007, 15, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Kane, J.R.; Miska, J.; Young, J.S.; Kanojia, D.; Kim, J.W.; Lesniak, M.S. Sui generis: Gene therapy and delivery systems for the treatment of glioblastoma. Neuro Oncol. 2015, 17 (Suppl. 2), ii24–ii36. [Google Scholar] [CrossRef] [PubMed]
- Boucher, P.D.; Im, M.M.; Freytag, S.O.; Shewach, D.S. A novel mechanism of synergistic cytotoxicity with 5-fluorocytosine and ganciclovir in double suicide gene therapy. Cancer Res. 2006, 66, 3230–3237. [Google Scholar] [CrossRef] [PubMed]
- Moriuchi, S.; Wolfe, D.; Tamura, M.; Yoshimine, T.; Miura, F.; Cohen, J.B.; Glorioso, J.C. Double suicide gene therapy using a replication defective herpes simplex virus vector reveals reciprocal interference in a malignant glioma model. Gene. Ther. 2002, 9, 584–591. [Google Scholar] [CrossRef] [PubMed]
- Tomicic, M.T.; Thust, R.; Kaina, B. Ganciclovir-induced apoptosis in HSV-1 thymidine kinase expressing cells: Critical role of DNA breaks, Bcl-2 decline and caspase-9 activation. Oncogene 2002, 21, 2141–2153. [Google Scholar] [CrossRef]
- Ichikawa, T.; Tamiya, T.; Adachi, Y.; Ono, Y.; Matsumoto, K.; Furuta, T.; Yoshida, Y.; Hamada, H.; Ohmoto, T. In vivo efficacy and toxicity of 5-fluorocytosine/cytosine deaminase gene therapy for malignant gliomas mediated by adenovirus. Cancer Gene. Ther. 2000, 7, 74–82. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Backus, H.H.; Wouters, D.; Ferreira, C.G.; van Houten, V.M.; Brakenhoff, R.H.; Pinedo, H.M.; Peters, G.J. Thymidylate synthase inhibition triggers apoptosis via caspases-8 and -9 in both wild-type and mutant p53 colon cancer cell lines. Eur. J. Cancer 2003, 39, 1310–1317. [Google Scholar] [CrossRef]
- Cloughesy, T.F.; Landolfi, J.; Vogelbaum, M.A.; Ostertag, D.; Elder, J.B.; Bloomfield, S.; Carter, B.; Chen, C.C.; Kalkanis, S.N.; Kesari, S.; et al. Durable complete responses in some recurrent high-grade glioma patients treated with Toca 511 + Toca FC. Neuro Oncol. 2018, 20, 1383–1392. [Google Scholar] [CrossRef] [PubMed]
- Philbrick, B.D.; Adamson, D.C. Early clinical trials of Toca 511 and Toca FC show a promising novel treatment for recurrent malignant glioma. Expert Opin. Investig. Drugs 2019, 28, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Twitty, C.G.; Diago, O.R.; Hogan, D.J.; Burrascano, C.; Ibanez, C.E.; Jolly, D.J.; Ostertag, D. Retroviral Replicating Vectors Deliver Cytosine Deaminase Leading to Targeted 5-Fluorouracil-Mediated Cytotoxicity in Multiple Human Cancer Types. Hum. Gene. Ther. Methods 2016, 27, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowska, A.; Nandhu, M.S.; Behera, P.; Chiocca, E.A.; Viapiano, M.S. Strategies in gene therapy for glioblastoma. Cancers 2013, 5, 1271–1305. [Google Scholar] [CrossRef] [PubMed]
- Saga, Y.; Suzuki, M.; Mizukami, H.; Kohno, T.; Takei, Y.; Fukushima, M.; Ozawa, K. Overexpression of thymidylate synthase mediates desensitization for 5-fluorouracil of tumor cells. Int. J. Cancer 2003, 106, 324–326. [Google Scholar] [CrossRef]
- Deschamps, M.; Mercier-Lethondal, P.; Certoux, J.M.; Henry, C.; Lioure, B.; Pagneux, C.; Cahn, J.Y.; Deconinck, E.; Robinet, E.; Tiberghien, P.; et al. Deletions within the HSV-tk transgene in long-lasting circulating gene-modified T cells infused with a hematopoietic graft. Blood 2007, 110, 3842–3852. [Google Scholar] [CrossRef]
- Ardiani, A.; Sanchez-Bonilla, M.; Black, M.E. Fusion enzymes containing HSV-1 thymidine kinase mutants and guanylate kinase enhance prodrug sensitivity in vitro and in vivo. Cancer Gene. Ther. 2010, 17, 86–96. [Google Scholar] [CrossRef]
- Wei, S.J.; Chao, Y.; Hung, Y.M.; Lin, W.C.; Yang, D.M.; Shih, Y.L.; Chang, L.Y.; Whang-Peng, J.; Yang, W.K. S- and G2-phase cell cycle arrests and apoptosis induced by ganciclovir in murine melanoma cells transduced with herpes simplex virus thymidine kinase. Exp. Cell Res. 1998, 241, 66–75. [Google Scholar] [CrossRef]
- Lumniczky, K.; Safrany, G. Cancer gene therapy: Combination with radiation therapy and the role of bystander cell killing in the anti-tumor effect. Pathol. Oncol. Res. 2006, 12, 118–124. [Google Scholar] [CrossRef]
- Wu, D.H.; Liu, L.; Chen, L.H. Antitumor effects and radiosensitization of cytosine deaminase and thymidine kinase fusion suicide gene on colorectal carcinoma cells. World J. Gastroenterol. 2005, 11, 3051–3055. [Google Scholar] [CrossRef]
- Rogulski, K.R.; Wing, M.S.; Paielli, D.L.; Gilbert, J.D.; Kim, J.H.; Freytag, S.O. Double suicide gene therapy augments the antitumor activity of a replication-competent lytic adenovirus through enhanced cytotoxicity and radiosensitization. Hum. Gene. Ther. 2000, 11, 67–76. [Google Scholar] [CrossRef]
- Chatterjee, R.; Yuan, L. Directed evolution of metabolic pathways. Trends Biotechnol. 2006, 24, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Fischer, U.; Steffens, S.; Frank, S.; Rainov, N.G.; Schulze-Osthoff, K.; Kramm, C.M. Mechanisms of thymidine kinase/ganciclovir and cytosine deaminase/5-fluorocytosine suicide gene therapy-induced cell death in glioma cells. Oncogene 2005, 24, 1231–1243. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.Y.; Huang, Z.H.; Lin, L.J.; Li, Z.; Yu, J.L.; Song, H.J.; Qian, Y.; Che, X.Y. Kinase domain insert containing receptor promoter controlled suicide gene system selectively kills human umbilical vein endothelial cells. World J. Gastroenterol. 2006, 12, 5331–5335. [Google Scholar] [CrossRef] [PubMed]
- Aghi, M.; Kramm, C.M.; Chou, T.C.; Breakefield, X.O.; Chiocca, E.A. Synergistic anticancer effects of ganciclovir/thymidine kinase and 5-fluorocytosine/cytosine deaminase gene therapies. J. Natl. Cancer Inst. 1998, 90, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Mei, L.; Wang, Y.; Liu, L.; Che, G. Double suicide genes selectively kill human umbilical vein endothelial cells. Virol. J. 2011, 8, 74. [Google Scholar] [CrossRef] [PubMed]
- Kubo, S.; Takagi-Kimura, M.; Tagawa, M.; Kasahara, N. Dual-vector prodrug activator gene therapy using retroviral replicating vectors. Cancer Gene. Ther. 2019, 26, 128–135. [Google Scholar] [CrossRef]
- Uckert, W.; Kammertons, T.; Haack, K.; Qin, Z.; Gebert, J.; Schendel, D.J.; Blankenstein, T. Double suicide gene (cytosine deaminase and herpes simplex virus thymidine kinase) but not single gene transfer allows reliable elimination of tumor cells in vivo. Hum. Gene. Ther. 1998, 9, 855–865. [Google Scholar] [CrossRef]
- Okura, H.; Smith, C.A.; Rutka, J.T. Gene therapy for malignant glioma. Mol. Cell Ther. 2014, 2, 21. [Google Scholar] [CrossRef] [PubMed]
- Logg, C.R.; Logg, A.; Tai, C.K.; Cannon, P.M.; Kasahara, N. Genomic stability of murine leukemia viruses containing insertions at the Env-3’ untranslated region boundary. J. Virol. 2001, 75, 6989–6998. [Google Scholar] [CrossRef]
- Yu, J.H.; Schaffer, D.V. Advanced targeting strategies for murine retroviral and adeno-associated viral vectors. Adv. Biochem. Eng. Biotechnol. 2005, 99, 147–167. [Google Scholar] [PubMed]
- Juratli, T.A.; Schackert, G.; Krex, D. Current status of local therapy in malignant gliomas—A clinical review of three selected approaches. Pharmacol. Ther. 2013, 139, 341–358. [Google Scholar] [CrossRef] [PubMed]
- Nehls, M.; Pfeifer, D.; Schorpp, M.; Hedrich, H.; Boehm, T. New member of the winged-helix protein family disrupted in mouse and rat nude mutations. Nature 1994, 372, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.; Mignot, G.; Chalmin, F.; Ladoire, S.; Bruchard, M.; Chevriaux, A.; Martin, F.; Apetoh, L.; Rebe, C.; Ghiringhelli, F. 5-Fluorouracil selectively kills tumor-associated myeloid-derived suppressor cells resulting in enhanced T cell-dependent antitumor immunity. Cancer Res. 2010, 70, 3052–3061. [Google Scholar] [CrossRef] [PubMed]
- Massey, A.J. Multiparametric Cell Cycle Analysis Using the Operetta High-Content Imager and Harmony Software with PhenoLOGIC. PLoS ONE 2015, 10, e0134306. [Google Scholar] [CrossRef] [PubMed]
- Wolf, D.; Goff, S.P. Host restriction factors blocking retroviral replication. Annu. Rev. Genet. 2008, 42, 143–163. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, A.V.; Sandoval, S. Pseudotyped Lentiviral Vectors: One Vector, Many Guises. Hum. Gene. Ther. Methods 2017, 28, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, Q.; Huang, W.; Li, X.; Wang, Y. Current status on the development of pseudoviruses for enveloped viruses. Rev. Med. Virol. 2018, 28, e1963. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, H.; Yao, C.; Su, F.; Guan, W.; Yan, S.; Ni, Z. Antitumor activity of combined endostatin and thymidine kinase gene therapy in C6 glioma models. Cancer Med. 2016, 5, 2477–2486. [Google Scholar] [CrossRef]
- Lee, J.Y.; Lee, D.H.; Kim, H.A.; Choi, S.A.; Lee, H.J.; Park, C.K.; Phi, J.H.; Wang, K.C.; Kim, S.U.; Kim, S.K. Double suicide gene therapy using human neural stem cells against glioblastoma: Double safety measures. J. Neurooncol. 2014, 116, 49–57. [Google Scholar] [CrossRef]
- Schiffer, D.; Annovazzi, L.; Casalone, C.; Corona, C.; Mellai, M. Glioblastoma: Microenvironment and Niche Concept. Cancers 2018, 11, 5. [Google Scholar] [CrossRef]
- Mitchell, L.A.; Lopez Espinoza, F.; Mendoza, D.; Kato, Y.; Inagaki, A.; Hiraoka, K.; Kasahara, N.; Gruber, H.E.; Jolly, D.J.; Robbins, J.M. Toca 511 gene transfer and treatment with the prodrug, 5-fluorocytosine, promotes durable antitumor immunity in a mouse glioma model. Neuro Oncol. 2017, 19, 930–939. [Google Scholar] [CrossRef] [PubMed]
- Ilkow, C.S.; Swift, S.L.; Bell, J.C.; Diallo, J.S. From scourge to cure: Tumour-selective viral pathogenesis as a new strategy against cancer. PLoS Pathog. 2014, 10, e1003836. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Balasenthil, S.; Reuther, J.; Frayna, A.; Wang, Y.; Chandler, D.S.; Abruzzo, L.V.; Rashid, A.; Rodriguez, J.; Lozano, G.; et al. DEAR1 is a chromosome 1p35 tumor suppressor and master regulator of TGF-beta-driven epithelial-mesenchymal transition. Cancer Discov. 2013, 3, 1172–1189. [Google Scholar] [CrossRef] [PubMed]
- Cloughesy, T.F.; Landolfi, J.; Hogan, D.J.; Bloomfield, S.; Carter, B.; Chen, C.C.; Elder, J.B.; Kalkanis, S.N.; Kesari, S.; Lai, A.; et al. Phase 1 trial of vocimagene amiretrorepvec and 5-fluorocytosine for recurrent high-grade glioma. Sci. Transl. Med. 2016, 8, 341ra375. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, M. The Par3/Par6/aPKC complex and epithelial cell polarity. Exp. Cell Res. 2013, 319, 1357–1364. [Google Scholar] [CrossRef]
- Van Roy, F. Beyond E-cadherin: Roles of other cadherin superfamily members in cancer. Nat. Rev. Cancer 2014, 14, 121–134. [Google Scholar] [CrossRef]
- Anastas, J.N.; Moon, R.T. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer 2013, 13, 11–26. [Google Scholar] [CrossRef]
- Liu, S.Y.; Aliyari, R.; Chikere, K.; Li, G.; Marsden, M.D.; Smith, J.K.; Pernet, O.; Guo, H.; Nusbaum, R.; Zack, J.A.; et al. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity 2013, 38, 92–105. [Google Scholar] [CrossRef]
- Ostertag, D.; Amundson, K.K.; Lopez Espinoza, F.; Martin, B.; Buckley, T.; Galvao da Silva, A.P.; Lin, A.H.; Valenta, D.T.; Perez, O.D.; Ibanez, C.E.; et al. Brain tumor eradication and prolonged survival from intratumoral conversion of 5-fluorocytosine to 5-fluorouracil using a nonlytic retroviral replicating vector. Neuro Oncol. 2012, 14, 145–159. [Google Scholar] [CrossRef]
- Yagiz, K.; Huang, T.T.; Lopez Espinoza, F.; Mendoza, D.; Ibanez, C.E.; Gruber, H.E.; Jolly, D.J.; Robbins, J.M. Toca 511 plus 5-fluorocytosine in combination with lomustine shows chemotoxic and immunotherapeutic activity with no additive toxicity in rodent glioblastoma models. Neuro Oncol. 2016, 18, 1390–1401. [Google Scholar] [CrossRef]
- Hiraoka, K.; Inagaki, A.; Kato, Y.; Huang, T.T.; Mitchell, L.A.; Kamijima, S.; Takahashi, M.; Matsumoto, H.; Hacke, K.; Kruse, C.A.; et al. Retroviral replicating vector-mediated gene therapy achieves long-term control of tumor recurrence and leads to durable anticancer immunity. Neuro Oncol. 2017, 19, 918–929. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.T.; Parab, S.; Burnett, R.; Diago, O.; Ostertag, D.; Hofman, F.M.; Espinoza, F.L.; Martin, B.; Ibanez, C.E.; Kasahara, N.; et al. Intravenous administration of retroviral replicating vector, Toca 511, demonstrates therapeutic efficacy in orthotopic immune-competent mouse glioma model. Hum. Gene. Ther. 2015, 26, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.; Eller, M.; Teigler, J.E.; Maloveste, S.M.; Schultz, B.T.; Soghoian, D.Z.; Lu, R.; Oster, A.F.; Chenine, A.L.; Alter, G.; et al. Cooperativity of HIV-Specific Cytolytic CD4 T Cells and CD8 T Cells in Control of HIV Viremia. J. Virol. 2015, 89, 7494–7505. [Google Scholar] [CrossRef] [PubMed]
- De La Rochere, P.; Guil-Luna, S.; Decaudin, D.; Azar, G.; Sidhu, S.S.; Piaggio, E. Humanized Mice for the Study of Immuno-Oncology. Trends Immunol. 2018, 39, 748–763. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.J.; Tai, C.K.; Kasahara, N.; Chen, T.C. Highly efficient and tumor-restricted gene transfer to malignant gliomas by replication-competent retroviral vectors. Hum. Gene. Ther. 2003, 14, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Tai, C.K.; Wang, W.J.; Chen, T.C.; Kasahara, N. Single-shot, multicycle suicide gene therapy by replication-competent retrovirus vectors achieves long-term survival benefit in experimental glioma. Mol. Ther. 2005, 12, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.T.; Hlavaty, J.; Ostertag, D.; Espinoza, F.L.; Martin, B.; Petznek, H.; Rodriguez-Aguirre, M.; Ibanez, C.E.; Kasahara, N.; Gunzburg, W.; et al. Toca 511 gene transfer and 5-fluorocytosine in combination with temozolomide demonstrates synergistic therapeutic efficacy in a temozolomide-sensitive glioblastoma model. Cancer Gene. Ther. 2013, 20, 544–551. [Google Scholar] [CrossRef]
- Takahashi, M.; Valdes, G.; Hiraoka, K.; Inagaki, A.; Kamijima, S.; Micewicz, E.; Gruber, H.E.; Robbins, J.M.; Jolly, D.J.; McBride, W.H.; et al. Radiosensitization of gliomas by intracellular generation of 5-fluorouracil potentiates prodrug activator gene therapy with a retroviral replicating vector. Cancer Gene. Ther. 2014, 21, 405–410. [Google Scholar] [CrossRef]
- De Melo, S.M.; Bittencourt, S.; Ferrazoli, E.G.; da Silva, C.S.; da Cunha, F.F.; da Silva, F.H.; Stilhano, R.S.; Denapoli, P.M.; Zanetti, B.F.; Martin, P.K.; et al. The Anti-Tumor Effects of Adipose Tissue Mesenchymal Stem Cell Transduced with HSV-Tk Gene on U-87-Driven Brain Tumor. PLoS ONE 2015, 10, e0128922. [Google Scholar] [CrossRef]
- Guadagno, E.; Presta, I.; Maisano, D.; Donato, A.; Pirrone, C.K.; Cardillo, G.; Corrado, S.D.; Mignogna, C.; Mancuso, T.; Donato, G.; et al. Role of Macrophages in Brain Tumor Growth and Progression. Int. J. Mol. Sci. 2018, 19, 1005. [Google Scholar] [CrossRef]
- Wu, T.D.; Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010, 26, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, I.H.; Cho, H.J.; Park, C.K.; Jung, Y.S.; Kim, Y.; Nam, S.H.; Kim, B.S.; Johnson, M.D.; Kong, D.S.; et al. Spatiotemporal Evolution of the Primary Glioblastoma Genome. Cancer Cell 2015, 28, 318–328. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, M.; Kim, Y.-S.; Lee, K.; Kang, M.; Shin, H.; Oh, J.-W.; Koo, H.; Kim, D.; Kim, Y.; Kong, D.-S.; et al. Novel Semi-Replicative Retroviral Vector Mediated Double Suicide Gene Transfer Enhances Antitumor Effects in Patient-Derived Glioblastoma Models. Cancers 2019, 11, 1090. https://doi.org/10.3390/cancers11081090
Lee M, Kim Y-S, Lee K, Kang M, Shin H, Oh J-W, Koo H, Kim D, Kim Y, Kong D-S, et al. Novel Semi-Replicative Retroviral Vector Mediated Double Suicide Gene Transfer Enhances Antitumor Effects in Patient-Derived Glioblastoma Models. Cancers. 2019; 11(8):1090. https://doi.org/10.3390/cancers11081090
Chicago/Turabian StyleLee, Mijeong, Yeon-Soo Kim, Kyoungmin Lee, Moonkyung Kang, Hyemi Shin, Jeong-Woo Oh, Harim Koo, Donggeon Kim, Yejin Kim, Doo-Sik Kong, and et al. 2019. "Novel Semi-Replicative Retroviral Vector Mediated Double Suicide Gene Transfer Enhances Antitumor Effects in Patient-Derived Glioblastoma Models" Cancers 11, no. 8: 1090. https://doi.org/10.3390/cancers11081090
APA StyleLee, M., Kim, Y.-S., Lee, K., Kang, M., Shin, H., Oh, J.-W., Koo, H., Kim, D., Kim, Y., Kong, D.-S., Nam, D.-H., & Lee, H. W. (2019). Novel Semi-Replicative Retroviral Vector Mediated Double Suicide Gene Transfer Enhances Antitumor Effects in Patient-Derived Glioblastoma Models. Cancers, 11(8), 1090. https://doi.org/10.3390/cancers11081090





