Novel Curcumin Inspired Bis-Chalcone Promotes Endoplasmic Reticulum Stress and Glioblastoma Neurosphere Cell Death
Abstract
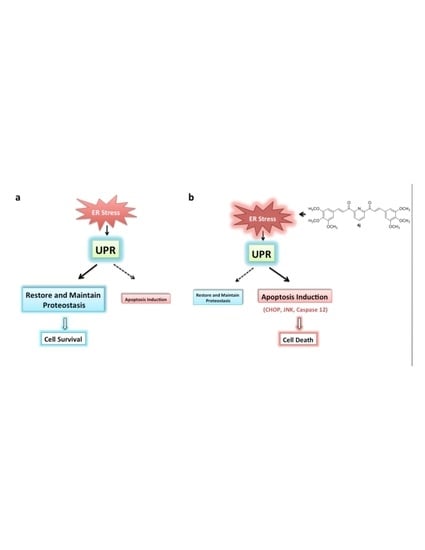
:1. Introduction
2. Results
2.1. Bis-Chalcone Synthesis
2.2. Bis-Chalcones Reduce Viability in GSCs
2.3. Bis-Chalcones 4c, 4h, 4j and 4n Substantially Reduce Viability in Six GSC Lines
2.4. Bis-Chalcones Reduce Neurosphere Formation at Sub Cytotoxic Levels
2.5. Bis-Chalcone 4j Does not Substantially Reduce p-Stat Activity
2.6. Bis-Chalcone 4j Induces Gene Expression Changes Consistent with ER Stress and UPR
2.6.1. Bis-Chalcones 4j Induces a Greater Transcriptional Impact Compared to 4n or Curcumin
2.6.2. Bis-Chalcone 4j Induces a Transcriptional Signature Consistent with ER Stress
2.6.3. Bis-Chalcone 4j Induce Similar Transcriptional Responses only in the Neurosphere Cell Lines
2.6.4. Drug Connectivity Analysis of 4j Supports ER Perturbation of Neurosphere Cells
2.7. Bis-Chalcone 4j Induces Robust Expression of CHOP and Promotes JNK and Caspase 12 Activity
2.8. Bis-Chalcone 4j Demonstrates Reduced Toxicity to Non-Cancer Stem Cells
3. Discussion
4. Materials and Methods
4.1. Chemistry
4.2. Cell Culture
4.3. Drug Treatment
4.4. Neurosphere Forming Assay
4.5. RNA Analysis
4.5.1. Transcriptional Impact
4.5.2. Functional Enrichment Analysis
4.5.3. CLUE Analysis
4.6. Western Blot Assay
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ostrom, Q.T.; Gittleman, H.; Truitt, G.; Boscia, A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro-Oncology 2018, 20 (Suppl. 4), iv1–iv86. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Chen, J.; Li, Y.; Yu, T.S.; McKay, R.M.; Burns, D.K.; Kernie, S.G.; Parada, L.F. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 2012, 488, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Kalkan, R. Glioblastoma Stem Cells as a New Therapeutic Target for Glioblastoma. Clin. Med. Insights Oncol. 2015, 9, 95–103. [Google Scholar] [CrossRef]
- Madden, E.; Logue, S.E.; Healy, S.J.; Manie, S.; Samali, A. The role of the unfolded protein response in cancer progression: From oncogenesis to chemoresistance. Biol. Cell 2019, 111, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Ciavattini, A.; Delli Carpini, G.; Serri, M.; Tozzi, A.; Leoni, F.; Di Loreto, E.; Saccucci, F. Unfolded protein response, a link between endometrioid ovarian carcinoma and endometriosis: A pilot study. Oncol. Lett. 2018, 16, 5449–5454. [Google Scholar] [CrossRef]
- Obacz, J.; Avril, T.; Le Reste, P.J.; Urra, H.; Quillien, V.; Hetz, C.; Chevet, E. Endoplasmic reticulum proteostasis in glioblastoma-From molecular mechanisms to therapeutic perspectives. Sci. Signal. 2017, 10. [Google Scholar] [CrossRef]
- Obacz, J.; Avril, T.; Rubio-Patino, C.; Bossowski, J.P.; Igbaria, A.; Ricci, J.E.; Chevet, E. Regulation of tumor-stroma interactions by the unfolded protein response. FEBS J. 2017. [Google Scholar] [CrossRef]
- Penaranda Fajardo, N.M.; Meijer, C.; Kruyt, F.A. The endoplasmic reticulum stress/unfolded protein response in gliomagenesis, tumor progression and as a therapeutic target in glioblastoma. Biochem. Pharmacol. 2016, 118, 1–8. [Google Scholar] [CrossRef]
- Mann, J. Natural products in cancer chemotherapy: Past, present and future. Nat. Rev. Cancer 2002, 2, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, G.A.; Shah, A.H.; Gersey, Z.C.; Shah, S.S.; Bregy, A.; Komotar, R.J.; Graham, R.M. Investigating the therapeutic role and molecular biology of curcumin as a treatment for glioblastoma. Ther. Adv. Med. Oncol. 2016, 8, 248–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Yu, J.; Cui, R.; Lin, J.; Ding, X. Curcumin in Treating Breast Cancer: A Review. J. Lab. Autom. 2016, 21, 723–731. [Google Scholar] [CrossRef] [PubMed]
- Mehta, H.J.; Patel, V.; Sadikot, R.T. Curcumin and lung cancer—A review. Targeted Oncol. 2014, 9, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, T. Targeting cancer stem cells by curcumin and clinical applications. Cancer Lett. 2014, 346, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Gersey, Z.C.; Rodriguez, G.A.; Barbarite, E.; Sanchez, A.; Walters, W.M.; Ohaeto, K.C.; Komotar, R.J.; Graham, R.M. Curcumin decreases malignant characteristics of glioblastoma stem cells via induction of reactive oxygen species. BMC Cancer 2017, 17, 99. [Google Scholar] [CrossRef]
- Cheng, A.L.; Hsu, C.H.; Lin, J.K.; Hsu, M.M.; Ho, Y.F.; Shen, T.S.; Ko, J.Y.; Lin, J.T.; Lin, B.R.; Ming-Shiang, W.; et al. Phase I clinical trial of curcumin, a chemopreventive agent, in patients with high-risk or pre-malignant lesions. Anticancer Res. 2001, 21, 2895–2900. [Google Scholar]
- Ramalingam, P.; Ko, Y.T. Enhanced oral delivery of curcumin from N-trimethyl chitosan surface-modified solid lipid nanoparticles: Pharmacokinetic and brain distribution evaluations. Pharm. Res. 2015, 32, 389–402. [Google Scholar] [CrossRef]
- Zhongfa, L.; Chiu, M.; Wang, J.; Chen, W.; Yen, W.; Fan-Havard, P.; Yee, L.D.; Chan, K.K. Enhancement of curcumin oral absorption and pharmacokinetics of curcuminoids and curcumin metabolites in mice. Cancer Chemother. Pharmacol. 2012, 69, 679–689. [Google Scholar] [CrossRef]
- Sasaki, H.; Sunagawa, Y.; Takahashi, K.; Imaizumi, A.; Fukuda, H.; Hashimoto, T.; Wada, H.; Katanasaka, Y.; Kakeya, H.; Fujita, M.; et al. Innovative preparation of curcumin for improved oral bioavailability. Biol. Pharm. Bull. 2011, 34, 660–665. [Google Scholar] [CrossRef]
- Chen, P.H.; Chang, C.K.; Shih, C.M.; Cheng, C.H.; Lin, C.W.; Lee, C.C.; Liu, A.J.; Ho, K.H.; Chen, K.C. The miR-204-3p-targeted IGFBP2 pathway is involved in xanthohumol-induced glioma cell apoptotic death. Neuropharmacology 2016, 110 (Pt A), 362–375. [Google Scholar] [CrossRef]
- Champelovier, P.; Chauchet, X.; Hazane-Puch, F.; Vergnaud, S.; Garrel, C.; Laporte, F.; Boutonnat, J.; Boumendjel, A. Cellular and molecular mechanisms activating the cell death processes by chalcones: Critical structural effects. Toxicol. In Vitro Int. J. Publ. Assoc. BIBRA 2013, 27, 2305–2315. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.W.; Overmeyer, J.H.; Young, A.M.; Erhardt, P.W.; Maltese, W.A. Synthesis and evaluation of indole-based chalcones as inducers of methuosis, a novel type of nonapoptotic cell death. J. Med. Chem. 2012, 55, 1940–1956. [Google Scholar] [CrossRef] [PubMed]
- Winter, E.; Devantier Neuenfeldt, P.; Chiaradia-Delatorre, L.D.; Gauthier, C.; Yunes, R.A.; Nunes, R.J.; Creczynski-Pasa, T.B.; Di Pietro, A. Symmetric bis-chalcones as a new type of breast cancer resistance protein inhibitors with a mechanism different from that of chromones. J. Med. Chem. 2014, 57, 2930–2941. [Google Scholar] [CrossRef]
- Lee, J.; Kotliarova, S.; Kotliarov, Y.; Li, A.; Su, Q.; Donin, N.M.; Pastorino, S.; Purow, B.W.; Christopher, N.; Zhang, W.; et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006, 9, 391–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Omran, F.; Al-Awadi, N.; Edun, M. Corrigendum-Synthesis of New 2-Pyrazoline Derivatives from 2, 6-Dicinnamoylpyridine and 1, 3-Dicinnamoylbenzene. J. Chem. Res.-Part S Synop. 1994, 333, 168–169. [Google Scholar]
- Reddy, D.B.; Seshamma, T.; Seenaiah, B.; Reddy, M.R. Synthesis and Biological Activity of Some New Bis (2-pyrazolin-3-yl) benzenes and-pyridines. Indian J. Chem. 1991, 30B, 46–51. [Google Scholar] [CrossRef]
- Constable, E.C.; Figgemeier, E.; Hougen, I.A.; Housecroft, C.E.; Neuburger, M.; Schaffner, S.; Whall, L.A. Hairpin helicates: A missing link between double-helicates and trefoil knots. Dalton Trans. 2005, 1168–1175. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Zhang, Q.; Yu, J.; Zhao, X.; Tian, Y.; Cui, Y.; Hao, X.; Yang, Y.; Qian, G. Solvent effect on two-photon absorption (TPA) of three novel dyes with large TPA cross-section and red emission. Dyes Pigments 2013, 97, 58–64. [Google Scholar] [CrossRef]
- Weissenberger, J.; Priester, M.; Bernreuther, C.; Rakel, S.; Glatzel, M.; Seifert, V.; Kogel, D. Dietary curcumin attenuates glioma growth in a syngeneic mouse model by inhibition of the JAK1,2/STAT3 signaling pathway. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010, 16, 5781–5795. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452e1417. [Google Scholar] [CrossRef]
- Stathias, V.; Jermakowicz, A.M.; Maloof, M.E.; Forlin, M.; Walters, W.; Suter, R.K.; Durante, M.A.; Williams, S.L.; Harbour, J.W.; Volmar, C.H.; et al. Drug and disease signature integration identifies synergistic combinations in glioblastoma. Nat. Commun. 2018, 9, 5315. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Li, Y.; Guo, Y.; Tang, J.; Jiang, J.; Chen, Z. New insights into the roles of CHOP-induced apoptosis in ER stress. Acta Biochim. Biophys. Sin. 2014, 46, 629–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joo, H.; Lee, H.J.; Shin, E.A.; Kim, H.; Seo, K.H.; Baek, N.I.; Kim, B.; Kim, S.H. c-Jun N-terminal Kinase-Dependent Endoplasmic Reticulum Stress Pathway is Critically Involved in Arjunic Acid Induced Apoptosis in Non-Small Cell Lung Cancer Cells. Phytother. Res. 2016, 30, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.Y.; Li, P.P.; Jin, F.S.; Yao, C.; Zhang, G.H.; Zang, T.; Ai, X. Ursolic acid induces ER stress response to activate ASK1-JNK signaling and induce apoptosis in human bladder cancer T24 cells. Cell. Signal. 2013, 25, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Zhu, H.; Morishima, N.; Li, E.; Xu, J.; Yankner, B.A.; Yuan, J. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature 2000, 403, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Du, Z.; Wang, W.; Song, M.; Sanidad, K.; Sukamtoh, E.; Zheng, J.; Tian, L.; Xiao, H.; Liu, Z.; et al. Structure-Activity Relationship of Curcumin: Role of the Methoxy Group in Anti-inflammatory and Anticolitis Effects of Curcumin. J. Agric. Food Chem. 2017, 65, 4509–4515. [Google Scholar] [CrossRef]
- Indira Priyadarsini, K. Chemical and structural features influencing the biological activity of curcumin. Curr. Pharm. Des. 2013, 19, 2093–2100. [Google Scholar]
- Zhou, D.; Ding, N.; Zhao, S.; Li, D.; Van Doren, J.; Qian, Y.; Wei, X.; Zheng, X. Synthesis and evaluation of curcumin-related compounds containing inden-2-one for their effects on human cancer cells. Biol. Pharm. Bull. 2014, 37, 1977–1981. [Google Scholar] [CrossRef]
- Bi, K.; Nishihara, K.; Machleidt, T.; Hermanson, S.; Wang, J.; Sakamuru, S.; Huang, R.; Xia, M. Identification of known drugs targeting the endoplasmic reticulum stress response. Anal. Bioanal. Chem. 2015, 407, 5343–5351. [Google Scholar] [CrossRef]
- Trivedi, M.V.; Laurence, J.S.; Siahaan, T.J. The role of thiols and disulfides on protein stability. Curr. Protein Pept. Sci. 2009, 10, 614–625. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Thomas, B.; Sachdeva, R.; Arterburn, L.; Frye, L.; Hatcher, P.G.; Cornwell, D.G.; Ma, J. Mechanism of arylating quinone toxicity involving Michael adduct formation and induction of endoplasmic reticulum stress. Proc. Natl. Acad. Sci. USA 2006, 103, 3604–3609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.; Du, S.; Du, Y.; Ren, J.; Ying, G.; Yan, Z. Glutathione reductase mediates drug resistance in glioblastoma cells by regulating redox homeostasis. J. Neurochem. 2018, 144, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Jackson, P.A.; Widen, J.C.; Harki, D.A.; Brummond, K.M. Covalent Modifiers: A Chemical Perspective on the Reactivity of alpha,beta-Unsaturated Carbonyls with Thiols via Hetero-Michael Addition Reactions. J. Med. Chem. 2017, 60, 839–885. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Bai, M.; Wang, X.; Tan, C.; Zhang, D.; Chang, L.; Li, G.; Xie, L.; Su, J.; Wen, Y. Estrogen receptor antagonist fulvestrant inhibits proliferation and promotes apoptosis of prolactinoma cells by regulating the IRE1/XBP1 signaling pathway. Mol. Med. Rep. 2018, 18, 4037–4041. [Google Scholar] [CrossRef] [PubMed]
- Minchenko, D.O.; Riabovol, O.O.; Ratushna, O.O.; Minchenko, O.H. Hypoxic regulation of the expression of genes encoded estrogen related proteins in U87 glioma cells: Effect of IRE1 inhibition. Endocr. Regul. 2017, 51, 8–19. [Google Scholar] [PubMed]
- Lhomond, S.; Avril, T.; Dejeans, N.; Voutetakis, K.; Doultsinos, D.; McMahon, M.; Pineau, R.; Obacz, J.; Papadodima, O.; Jouan, F.; et al. Dual IRE1 RNase functions dictate glioblastoma development. EMBO Mol. Med. 2018, 10, e7929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jabouille, A.; Delugin, M.; Pineau, R.; Dubrac, A.; Soulet, F.; Lhomond, S.; Pallares-Lupon, N.; Prats, H.; Bikfalvi, A.; Chevet, E.; et al. Glioblastoma invasion and cooption depend on IRE1alpha endoribonuclease activity. Oncotarget 2015, 6, 24922–24934. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Wuts, P.G.; Greene, T.W. Greene’s Protective Groups in Organic Synthesis; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar]
- Graham, R.M.; Hernandez, F.; Puerta, N.; De Angulo, G.; Webster, K.A.; Vanni, S. Resveratrol augments ER stress and the cytotoxic effects of glycolytic inhibition in neuroblastoma by downregulating Akt in a mechanism independent of SIRT1. Exp. Mol. Med. 2016, 48, e210. [Google Scholar] [CrossRef]
- Hombach-Klonisch, S.; Mehrpour, M.; Shojaei, S.; Harlos, C.; Pitz, M.; Hamai, A.; Siemianowicz, K.; Likus, W.; Wiechec, E.; Toyota, B.D.; et al. Glioblastoma and chemoresistance to alkylating agents: Involvement of apoptosis, autophagy, and unfolded protein response. Pharmacol. Ther. 2018, 184, 13–41. [Google Scholar] [CrossRef] [PubMed]
- Dadey, D.Y.; Kapoor, V.; Khudanyan, A.; Urano, F.; Kim, A.H.; Thotala, D.; Hallahan, D.E. The ATF6 pathway of the ER stress response contributes to enhanced viability in glioblastoma. Oncotarget 2016, 7, 2080–2092. [Google Scholar] [CrossRef] [PubMed]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sansalone, L.; Veliz, E.A.; Myrthil, N.G.; Stathias, V.; Walters, W.; Torrens, I.I.; Schürer, S.C.; Vanni, S.; Leblanc, R.M.; Graham, R.M. Novel Curcumin Inspired Bis-Chalcone Promotes Endoplasmic Reticulum Stress and Glioblastoma Neurosphere Cell Death. Cancers 2019, 11, 357. https://doi.org/10.3390/cancers11030357
Sansalone L, Veliz EA, Myrthil NG, Stathias V, Walters W, Torrens II, Schürer SC, Vanni S, Leblanc RM, Graham RM. Novel Curcumin Inspired Bis-Chalcone Promotes Endoplasmic Reticulum Stress and Glioblastoma Neurosphere Cell Death. Cancers. 2019; 11(3):357. https://doi.org/10.3390/cancers11030357
Chicago/Turabian StyleSansalone, Lorenzo, Eduardo A. Veliz, Nadia G. Myrthil, Vasileios Stathias, Winston Walters, Ingrid I. Torrens, Stephan C. Schürer, Steven Vanni, Roger M. Leblanc, and Regina M. Graham. 2019. "Novel Curcumin Inspired Bis-Chalcone Promotes Endoplasmic Reticulum Stress and Glioblastoma Neurosphere Cell Death" Cancers 11, no. 3: 357. https://doi.org/10.3390/cancers11030357