Plasma Androgen Receptor in Prostate Cancer
Abstract
:1. Introduction
2. Clinical Prognostic Factors in Prostate Cancer
3. Molecular Factors in Driving Castration-Resistant Prostate Cancer (CRPC) Emergence
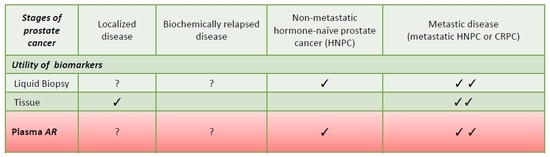
4. Biomarker Tools in Prostate Cancer
5. Detection of Androgen Receptor in Plasma
6. Plasma Androgen Receptor (AR) and Hormonal Treatments
7. Plasma Androgen Receptor (AR) and Chemotherapy
8. Plasma Androgen Receptor (AR) and Novel Drugs
9. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer. J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Davies, A.; Conteduca, V.; Zoubeidi, A.; Beltran, H. Biological evolution of castration-resistant prostate cancer. Eur. Urol. Focus. 2019, 5, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Scher, H.I.; Halabi, S.; Tannock, I.; Morris, M.; Sternberg, C.N.; Carducci, M.A.; Eisenberger, M.A.; Higano, C.; Bubley, G.J.; Dreicer, R.; et al. Design and endpoints of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: Recommendations of the prostate cancer clinical trials working group. J. Clin. Oncol. 2008, 26, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Scher, H.I.; Morris, M.J.; Stadler, W.M.; Higano, C.; Basch, E.; Fizazi, K.; Antonarakis, E.S.; Beer, T.M.; Carducci, M.A.; Chi, K.N.; et al. Trial design and objectives for castration-resistant prostate cancer: Updated recommendations from the prostate cancer clinical trials working group 3. J. Clin. Oncol. 2016, 34, 1402–1418. [Google Scholar] [CrossRef]
- Halabi, S.; Lin, C.Y.; Kelly, W.K.; Fizazi, K.S.; Moul, J.W.; Kaplan, E.B.; Morris, M.J.; Small, E.J. Updated prognostic model for predicting overall survival in first-line chemotherapy for patients with metastatic castration-resistant prostate cancer. J. Clin. Oncol. 2014, 32, 671–677. [Google Scholar] [CrossRef]
- Gleason, D.F.; Mellinger, G.T. Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. J. Urol. 1974, 111, 58–64. [Google Scholar] [CrossRef]
- Catalona, W.J.; Richie, J.P.; Ahmann, F.R.; Hudson, M.A.; Scardino, P.T.; Flanigan, R.C.; DeKernion, J.B.; Ratliff, T.L.; Kavoussi, L.R.; Dalkin, B.L.; et al. Comparison of digital rectal examination and serum prostate specific antigen in the early detection of prostate cancer: Results of a multicenter clinical trial of 6, 630 men. J. Urol. 1994, 151, 1283–1290. [Google Scholar] [CrossRef]
- Sobin, L.H.; Gospodarowicz, M.; Wittekind, C. TNM Classification of Malignant Tumors, 7th ed.; UICC International Union Against Cancer; Wiley-Blackwell: New York, NY, USA, 2009. [Google Scholar]
- Oderda, M.; Cozzi, G.; Daniele, L.; Sapino, A.; Munegato, S.; Renne, G.; De Cobelli, O.; Gontero, P. Cell-cycle progression-score might improve the current risk assessment in newly diagnosed prostate cancer patients. Urology 2017, 102, 73–78. [Google Scholar] [CrossRef]
- Brand, T.C.; Zhang, N.; Crager, M.R.; Maddala, T.; Dee, A.; Sesterhenn, I.A.; Simko, J.P.; Cooperberg, M.R.; Srivastava, S.; Rosner, I.L.; et al. Patient-specific meta-analysis of 2 clinical validation studies to predict pathologic outcomes in prostate cancer using the 17-gene genomic prostate score. Urology 2016, 89, 69–75. [Google Scholar] [CrossRef]
- Erho, N.; Crisan, A.; Vergara, I.A.; Mitra, A.P.; Ghadessi, M.; Buerki, C.; Bergstralh, E.J.; Kollmeyer, T.; Fink, S.; Haddad, Z.; et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS ONE 2013, 8, e66855. [Google Scholar] [CrossRef]
- Mateo, J.; Fizazi, K.; Gillessen, S.; Heidenreich, A.; Perez-Lopez, R.; Oyen, W.J.G.; Heidenreich, A.; Perez-Lopez, R.; Oyen, W.J.G.; Shore, N.; et al. Managing nonmetastatic castration-resistant prostate cancer. Eur. Urol. 2018, 75, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Francini, E.; Gray, K.P.; Xie, W.; Shaw, G.K.; Valença, L.; Bernard, B.; Albiges, L.; Harshman, L.C.; Kantoff, P.W.; Taplin, M.E.; et al. Time of metastatic disease presentation and volume of disease are prognostic for metastatic hormone sensitive prostate cancer (mHSPC). Prostate 2018, 78, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, C.J.; Chen, Y.H.; Carducci, M.; Liu, G.; Jarrard, D.F.; Eisenberger, M.; Wong, Y.N.; Hahn, N.; Kohli, M.; Cooney, M.M.; et al. Chemohormonal therapy in metastatic hormone-sensitive prostate cancer. N. Eng. J. Med. 2015, 373, 737–746. [Google Scholar] [CrossRef] [PubMed]
- James, N.D.; Sydes, M.R.; Clarke, N.W.; Mason, M.D.; Dearnaley, D.P.; Spears, M.R.; Ritchie, A.W.; Parker, C.C.; Russell, J.M.; Attard, G.; et al. Addition of docetaxel, zoledronic acid, or both to first-line long-term hormone therapy in prostate cancer (STAMPEDE): Survival results from an adaptive, multiarm, multistage, platform randomised controlled trial. Lancet 2016, 387, 1163–1177. [Google Scholar] [CrossRef]
- Fizazi, K.; Tran, N.; Fein, L.; Matsubara, N.; Rodriguez-Antolin, A.; Alekseev, B.Y.; Özgüroğlu, M.; Ye, D.; Feyerabend, S.; Protheroe, A.; et al. Abiraterone plus prednisone in metastatic, castration-sensitive prostate cancer. N. Engl. J. Med. 2017, 377, 352–360. [Google Scholar] [CrossRef]
- Harshman, L.C.; Chen, Y.H.; Liu, G.; Carducci, M.A.; Jarrard, D.; Dreicer, R.; Hahn, N.; Garcia, J.A.; Hussain, M.; Shevrin, D.; et al. Seven-month prostate-specific antigen is prognostic in metastatic hormone-sensitive prostate cancer treated with androgen deprivation with or without docetaxel. J. Clin. Oncol. 2018, 36, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Chi, K.N.; Kheoh, T.; Ryan, C.J.; Molina, A.; Bellmunt, J.; Vogelzang, N.J.; Rathkopf, D.E.; Fizazi, K.; Kantoff, P.W.; Li, J.; et al. A prognostic index model for predicting overall survival in patients with metastatic castration-resistant prostate cancer treated with abiraterone acetate after docetaxel. Ann. Oncol. 2016, 27, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Halabi, S.; Small, E.J.; Kantoff, P.W.; Kattan, M.W.; Kaplan, E.B.; Dawson, N.A.; Levine, E.G.; Blumenstein, B.A.; Vogelzang, N.J. Prognostic model for predicting survival in men with hormone-refractory metastatic prostate cancer. J. Clin. Oncol. 2003, 21, 1232–1237. [Google Scholar] [CrossRef]
- Halabi, S.; Lin, C.Y.; Small, E.J.; Armstrong, A.J.; Kaplan, E.B.; Petrylak, D.; Sternberg, C.N.; Shen, L.; Oudard, S.; de Bono, J.; et al. Prognostic model predicting metastatic castration-resistant prostate cancer survival in men treated with second-line chemotherapy. J. Natl. Cancer Inst. 2013, 105, 1729–1737. [Google Scholar] [CrossRef]
- Ryan, C.J.; Smith, M.R.; de Bono, J.S.; Molina, A.; Logothetis, C.J.; de Souza, P.; Fizazi, K.; Mainwaring, P.; Piulats, J.M.; Ng, S.; et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N. Engl. J. Med. 2013, 368, 138–148. [Google Scholar] [CrossRef]
- Beer, T.M.; Armstrong, A.J.; Rathkopf, D.E.; Loriot, Y.; Sternberg, C.N.; Higano, C.S.; Iversen, P.; Bhattacharya, S.; Carles, J.; Chowdhury, S.; et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N. Engl. J. Med. 2014, 371, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.; Shah, S.P.; Chin, S.F. The genomic and transcriptomic architecture of 2000 breast tumors reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Beltran, H.; Wyatt, A.W.; Chedgy, E.C.; Donoghue, A.; Annala, M.; Warner, E.W.; Beja, K.; Sigouros, M.; Mo, F.; Fazli, L.; et al. Impact of therapy on genomics and transcriptomics in high-risk prostate cancer treated with neoadjuvant docetaxel and androgen deprivation therapy. Clin. Cancer Res. 2017, 23, 6802–6811. [Google Scholar] [CrossRef] [PubMed]
- Grasso, C.S.; Wu, Y.M.; Robinson, D.R.; Cao, X.; Dhanasekaran, S.M.; Khan, A.P.; Quist, M.J.; Jing, X.; Lonigro, R.J.; Brenner, J.C.; et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012, 487, 239–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyatt, A.W.; Gleave, M.E. Targeting the adaptive molecular landscape of castration-resistant prostate cancer. EMBO Mol. Med. 2015, 7, 878–894. [Google Scholar] [CrossRef]
- Mostaghel, E.A.; Nelson, P.S.; Lange, P.; Lin, D.W.; Taplin, M.E.; Balk, S.; Ellis, W.; Kantoff, P.; Marck, B.; Tamae, D.; et al. Targeted androgen pathway suppression in localized prostate cancer: A pilot study. J. Clin. Oncol. 2014, 32, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Taplin, M.E.; Montgomery, B.; Logothetis, C.J.; Bubley, G.J.; Richie, J.P.; Dalkin, B.L.; Sanda, M.G.; Davis, J.W.; Loda, M.; True, L.D.; et al. Intense androgen-deprivation therapy with abiraterone acetate plus leuprolide acetate in patients with localized high-risk prostate cancer: results of a randomized phase II neoadjuvant study. J. Clin. Oncol. 2014, 32, 3705–3715. [Google Scholar] [CrossRef]
- Montgomery, B.; Tretiakova, M.S.; Joshua, A.M.; Gleave, M.E.; Fleshner, N.; Bubley, G.J.; Mostaghel, E.A.; Chi, K.N.; Lin, D.W.; Sanda, M.; et al. Neoadjuvant enzalutamide prior to prostatectomy. Clin. Cancer Res. 2017, 23, 2169–2176. [Google Scholar] [CrossRef]
- Beltran, H.; Prandi, D.; Mosquera, J.M.; Benelli, M.; Puca, L.; Cyrta, J.; Marotz, C.; Giannopoulou, E.; Chakravarthi, B.V.; Varambally, S.; et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 2016, 22, 298–305. [Google Scholar] [CrossRef] [Green Version]
- Ku, S.Y.; Rosario, S.; Wang, Y.; Mu, P.; Seshadri, M.; Goodrich, Z.W.; Goodrich, M.M.; Labbé, D.P.; Gomez, E.C.; Wang, J.; et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017, 355, 78–83. [Google Scholar] [CrossRef]
- Zou, M.; Toivanen, R.; Mitrofanova, A.; Floch, N.; Hayati, S.; Sun, Y.; Le Magnen, C.; Chester, D.; Mostaghel, E.A.; Califano, A.; et al. Transdifferentiation as a mechanism of treatment resistance in a mouse model of castration-resistant prostate cancer. Cancer Discov. 2017, 7, 736–749. [Google Scholar] [CrossRef] [PubMed]
- Akamatsu, S.; Wyatt, A.W.; Lin, D.; Lysakowski, S.; Zhang, F.; Kim, S.; Tse, C.; Wang, K.; Mo, F.; Haegert, A.; et al. The placental gene PEG10 promotes progression of neuroendocrine prostate cancer. Cell Rep. 2015, 12, 922–936. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Wyatt, A.W.; Xue, H.; Wang, Y.; Dong, X.; Haegert, A.; Wu, R.; Brahmbhatt, S.; Mo, F.; Jong, L.; et al. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res. 2014, 74, 1272–1283. [Google Scholar] [CrossRef] [PubMed]
- Davies, A.H.; Beltran, H.; Zoubeidi, A. Cellular plasticity and the neuroendocrine phenotype in prostate cancer. Nat. Rev. Urol. 2018, 15, 271–286. [Google Scholar] [CrossRef] [PubMed]
- Sowalsky, A.G.; Ye, H.; Bhasin, M.; Van Allen, E.M.; Loda, M.; Lis, R.T.; Montaser-Kouhsari, L.; Calagua, C.; Ma, F.; Russo, J.W.; et al. Neoadjuvant-intensive androgen deprivation therapy selects for prostate tumor foci with diverse subclonal oncogenic alterations. Cancer Res. 2018, 78, 4716–4730. [Google Scholar] [CrossRef] [PubMed]
- Varambally, S.; Yu, J.; Laxman, B. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell 2005, 8, 393–406. [Google Scholar] [CrossRef] [Green Version]
- Tomlins, S.A.; Mehra, R.; Rhodes, D.R. Integrative molecular concept modeling of prostate cancer progression. Nat. Genet. 2007, 39, 41–51. [Google Scholar] [CrossRef]
- Taylor, B.S.; Schultz, N.; Hieronymus, H. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010, 18, 11–22. [Google Scholar] [CrossRef]
- Robinson, D.; Van Allen, E.M.; Wu, Y.M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.M.; Montgomery, B.; Taplin, M.E.; Pritchard, C.C.; Attard, G.; et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015, 162, 454. [Google Scholar] [CrossRef]
- Barbieri, C.E.; Baca, S.C.; Lawrence, M.S.; Demichelis, F.; Blattner, M.; Theurillat, J.P.; White, T.A.; Stojanov, P.; Van Allen, E.; Stransky, N.; et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012, 44, 685–689. [Google Scholar] [CrossRef] [Green Version]
- Espiritu, S.M.G.; Liu, L.Y.; Rubanova, Y.; Bhandari, V.; Holgersen, E.M.; Szyca, L.M.; Fox, N.S.; Chua, M.L.K.; Yamaguchi, T.N.; Heisler, L.E.; et al. The evolutionary landscape of localized prostate cancers drives clinical aggression. Cell 2018, 173, 1003–1013. [Google Scholar] [CrossRef] [PubMed]
- Boutros, P.C.; Fraser, M.; Harding, N.J.; de Borja, R.; Trudel, D.; Lalonde, E.; Meng, A.; Hennings-Yeomans, P.H.; McPherson, A.; Sabelnykova, V.Y.; et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat. Genet. 2015, 47, 736–745. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Wang, J.; Lampert, E.; Schlanger, S.; DePriest, A.D.; Hu, Q.; Gomez, E.C.; Murakam, M.; Glenn, S.T.; Conroy, J.; et al. Intratumoral and intertumoral genomic heterogeneity of multifocal localized prostate cancer impacts molecular classifications and genomic prognosticators. Eur. Urol. 2017, 71, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Cooper, C.S.; Eeles, R.; Wedge, D.C.; Van Loo, P.; Gundem, G.; Alexandrov, L.B.; Kremeyer, B.; Butler, A.; Lynch, A.G.; Camacho, N.; et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat. Genet. 2015, 47, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Beltran, H.; Eng, K.; Mosquera, J.M.; Sigaras, A.; Romanel, A.; Rennert, H.; Kossai, M.; Pauli, C.; Faltas, B.; Fontugne, J.; et al. Whole-exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncol. 2015, 1, 466–474. [Google Scholar] [CrossRef]
- Carreira, S.; Romanel, A.; Goodall, J.; Grist, E.; Ferraldeschi, R.; Miranda, S.; Prandi, D.; Lorente, D.; Frenel, J.S.; Pezaro, C.; et al. Tumor clone dynamics in lethal prostate cancer. Sci. Transl. Med. 2014, 6, 254ra125. [Google Scholar] [CrossRef]
- Conteduca, V.; Zamarchi, R.; Rossi, E.; Condelli, V.; Troiani, L.; Aieta, M. Circulating tumor cells: Utopia or reality? Future Oncol. 2013, 9, 1337–1352. [Google Scholar] [CrossRef]
- Danila, D.C.; Heller, G.; Gignac, G.A.; Gonzalez-Espinoza, R.; Anand, A.; Tanaka, E.; Lilja, H.; Schwartz, L.; Larson, S.; Fleisher, M.; et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin. Cancer Res. 2007, 13, 7053–7058. [Google Scholar] [CrossRef]
- Scher, H.I.; Jia, X.; de Bono, J.S.; Fleisher, M.; Pienta, K.J.; Raghavan, D.; Heller, G. Circulating tumor cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol. 2009, 10, 233–239. [Google Scholar] [CrossRef]
- Goldkorn, A.; Ely, B.; Quinn, D.I.; Tangen, C.M.; Fink, L.M.; Xu, T.; Twardowski, P.; Van Veldhuizen, P.J.; Agarwal, N.; Carducci, M.A.; et al. Circulating tumor cell counts are prognostic of overall survival in SWOG S0421: A phase III trial of docetaxel with or without atrasentan for metastatic castration-resistant prostate cancer. J. Clin. Oncol. 2014, 32, 1136–1142. [Google Scholar] [CrossRef]
- Scher, H.I.; Heller, G.; Molina, A.; Attard, G.; Danila, D.C.; Jia, X.; Peng, W.; Sandhu, S.K.; Olmos, D.; Riisnaes, R.; et al. Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J. Clin. Oncol. 2015, 33, 1348–1355. [Google Scholar] [CrossRef] [PubMed]
- Lorente, D.; Olmos, D.; Mateo, J.; Bianchini, D.; Seed, G.; Fleisher, M.; Danila, D.C.; Flohr, P.; Crespo, M.; Figueiredo, I.; et al. Decline in circulating tumor cell count and treatment outcome in advanced prostate cancer. Eur. Urol. 2016, 70, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Lorente, D.; Olmos, D.; Mateo, J.; Dolling, D.; Bianchini, D.; Seed, G.; Flohr, P.; Crespo, M.; Figueiredo, I.; Miranda, S.; et al. Circulating tumor cell increase as a biomarker of disease progression in metastatic castration-resistant prostate cancer patients with low baseline CTC counts. Ann. Oncol. 2018, 29, 1554–1560. [Google Scholar] [PubMed]
- Antonarakis, E.S.; Lu, C.; Wang, H.; Luber, B.; Nakazawa, M.; Roeser, J.C.; Chen, Y.; Mohammad, T.A.; Chen, Y.; Fedor, H.L.; et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014, 371, 1028–1038. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Lu, C.; Luber, B.; Wang, H.; Chen, Y.; Nakazawa, M.; Nadal, R.; Paller, C.J.; Denmeade, S.R.; Carducci, M.A.; et al. Androgen receptor splice variant 7 and efficacy of taxane chemotherapy in patients with metastatic castration-resistant prostate cancer. JAMA Oncol. 2015, 1, 582–591. [Google Scholar] [CrossRef]
- Scher, H.I.; Lu, D.; Schreiber, N.A.; Louw, J.; Graf, R.P.; Vargas, H.A.; Johnson, A.; Jendrisak, A.; Bambury, R.; Danila, D.; et al. Association of AR-V7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncol. 2016, 2, 1441–1449. [Google Scholar] [CrossRef]
- Scher, H.I.; Graf, R.P.; Schreiber, N.A.; Jayaram, A.; Winquist, E.; McLaughlin, B.; Lu, D.; Fleisher, M.; Orr, S.; Lowes, L.; et al. Assessment of the validity of nuclear-localized androgen receptor splice variant 7 in circulating tumor cells as a predictive biomarker for castration-resistant prostate cancer. JAMA Oncol. 2018, 4, 1179–1186. [Google Scholar] [CrossRef]
- Romanel, A.; Tandefelt, D.G.; Conteduca, V.; Jayaram, A.; Casiraghi, N.; Wetterskog, D.; Salvi, S.; Amadori, D.; Zafeiriou, Z.; Rescigno, P.; et al. Plasma AR and abiraterone-resistant prostate cancer. Sci. Transl. Med. 2015, 312, 312re10. [Google Scholar] [CrossRef]
- Salvi, S.; Casadio, V.; Conteduca, V.; Burgio, S.L.; Menna, C.; Bianchi, E.; Rossi, L.; Carretta, E.; Masini, C.; Amadori, D.; et al. Circulating cell-free AR and CYP17A1 copy number variations may associate with outcome of metastatic castration-resistant prostate cancer patients treated with abiraterone. Br. J. Cancer 2015, 112, 1717–1724. [Google Scholar] [CrossRef]
- Salvi, S.; Casadio, V.; Conteduca, V.; Lolli, C.; Gurioli, G.; Martignano, F.; Schepisi, G.; Testoni, S.; Scarpi, E.; Amadori, D.; et al. Circulating AR copy number and outcome to enzalutamide in docetaxel-treated metastatic castration-resistant prostate cancer. Oncotarget 2016, 7, 37839–37845. [Google Scholar] [CrossRef]
- Azad, A.A.; Volik, S.V.; Wyatt, A.W.; Haegert, A.; Le Bihan, S.; Bell, R.H.; Anderson, S.A.; McConeghy, B.; Shukin, R.; Bazov, J.; et al. Androgen receptor gene aberrations in circulating cell-free DNA: Biomarkers of therapeutic resistance in castration-resistant prostate cancer. Clin. Cancer Res. 2015, 21, 2315–2324. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, A.W.; Azad, A.A.; Volik, S.V.; Annala, M.; Beja, K.; McConeghy, B.; Haegert, A.; Warner, E.W.; Mo, F.; Brahmbhatt, S.; et al. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol. 2016, 2, 1598–1606. [Google Scholar] [CrossRef] [PubMed]
- Conteduca, V.; Wetterskog, D.; Sharabiani, M.T.A.; Grande, E.; Fernandez-Perez, M.P.; Jayaram, A.; Salvi, S.; Castellano, D.; Romanel, A.; Lolli, C.; et al. Androgen receptor gene status in plasma DNA associates with worse outcome on enzalutamide or abiraterone for castration-resistant prostate cancer: A multi-institution correlative biomarker study. Ann. Oncol. 2017, 28, 1508–1516. [Google Scholar] [CrossRef] [PubMed]
- Annala, M.; Vandekerkhove, G.; Khalaf, D.; Taavitsainen, S.; Beja, K.; Warner, E.W.; Sunderland, K.; Kollmannsberger, C.; Eigl, B.J.; Finch, D.; et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 2018, 8, 444–457. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, T.; Lejeune, P.; Köhr, S.; Neuhaus, R.; Faus, H.; Gelato, K.A.; Busemann, M.; Cleve, A.; Lücking, U.; von Nussbaum, F.; et al. BAY 1024767 blocks androgen receptor mutants found in castration-resistant prostate cancer patients. Oncotarget 2016, 7, 6015–6028. [Google Scholar] [CrossRef] [Green Version]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- de Bono, J.S.; Logothetis, C.J.; Molina, A.; Fizazi, K.; North, S.; Chu, L.; Chi, K.N.; Jones, R.J.; Goodman, O.B. Jr.; Saad, F.; et al. Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 2011, 364, 1995–2005. [Google Scholar] [CrossRef]
- Scher, H.I.; Fizazi, K.; Saad, F.; Taplin, M.E.; Sternberg, C.N.; Miller, K.; de Wit, R.; Mulders, P.; Chi, K.N.; Shore, N.D.; et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 2012, 367, 1187–1197. [Google Scholar] [CrossRef]
- Tannock, I.F.; de Wit, R.; Berry, W.R.; Horti, J.; Pluzanska, A.; Chi, K.N.; Oudard, S.; Théodore, C.; James, N.D.; Turesson, I.; et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N. Engl. J. Med. 2004, 351, 1502–1512. [Google Scholar] [CrossRef]
- de Bono, J.S.; Oudard, S.; Ozguroglu, M.; Hansen, S.; Machiels, J.P.; Kocak, I.; Gravis, G.; Bodrogi, I.; Mackenzie, M.J.; Shen, L.; et al. Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: A randomised open-label trial. Lancet 2010, 376, 1147–1154. [Google Scholar] [CrossRef]
- Mateo, J.; Carreira, S.; Sandhu, S.; Miranda, S.; Mossop, H.; Perez-Lopez, R.; Nava Rodrigues, D.; Robinson, D.; Omlin, A.; Tunariu, N.; et al. DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 2015, 373, 1697–1708. [Google Scholar] [CrossRef] [PubMed]
- de Bono, J.S.; De Giorgi, U.; Nava Rodrigues, D.; Massard, C.; Bracarda, S.; Font, A.; Arranz Arija, J.A.; Shih, K.C.; Radavoi, G.D.; Xu, N.; et al. Randomized Phase II Study of Akt blockade with or without ipatasertib in abiraterone-treated patients with metastatic prostate cancer with and without PTEN Loss. Clin. Cancer Res. 2018, 25, 928–936. [Google Scholar] [CrossRef] [PubMed]
- Chi, K.N.; Thomas, S.; Agarwal, N.; Feng, F.; Attard, G.; Wyatt, A.W.; Gormley, M.; Ricci, D.S.; Lopez-Gitlitz, A.; Deprince, K.; et al. Androgen receptor (AR) aberrations in patients (Pts) with metastatic castration-sensitive prostate cancer (mCSPC) treated with apalutamide (APA) plus androgen deprivation therapy (ADT) in TITAN. Ann. Oncol. 2019. [Google Scholar] [CrossRef]
- Watson, P.A.; Arora, V.K.; Sawyers, C.L. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer 2015, 15, 701–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beltran, H.; Yelensky, R.; Frampton, G.M.; Park, K.; Downing, S.R.; MacDonald, T.Y.; Jarosz, M.; Lipson, D.; Tagawa, S.T.; Nanus, D.M.; et al. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur. Urol. 2013, 63, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, K.; Uchida, J.; Nishino, K.; Kumagai, T.; Okuyama, T.; Okami, J.; Higashiyama, M.; Kodama, K.; Imamura, F.; Kato, K. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin. Cancer. Res. 2011, 17, 7808–7815. [Google Scholar] [CrossRef]
- Asim, M.; Massie, C.E.; Orafidiya, F.; Pértega-Gomes, N.; Warren, A.Y.; Esmaeili, M.; Selth, L.A.; Zecchini, H.I.; Luko, K.; Qureshi, A.; et al. Choline kinase alpha as an androgen receptor chaperone and prostate cancer therapeutic target. J. Natl. Cancer Inst. 2015, 108. [Google Scholar] [CrossRef]
- Conteduca, V.; Scarpi, E.; Caroli, P.; Salvi, S.; Lolli, C.; Burgio, S.L.; Menna, C.; Schepisi, G.; Testoni, S.; Gurioli, G.; et al. Circulating androgen receptor combined with 18F-fluorocholine PET/CT metabolic activity and outcome to androgen receptor signalling-directed therapies in castration-resistant prostate cancer. Sci. Rep. 2017, 7, 15541. [Google Scholar] [CrossRef]
- Conteduca, V.; Burgio, S.L.; Menna, C.; Carretta, E.; Rossi, L.; Bianchi, E.; Masini, C.; Amadori, D.; De Giorgi, U. Chromogranin A is a potential prognostic marker in prostate cancer patients treated with enzalutamide. Prostate 2014, 74, 1691–1696. [Google Scholar] [CrossRef]
- Burgio, S.L.; Conteduca, V.; Menna, C.; Carretta, E.; Rossi, L.; Bianchi, E.; Kopf, B.; Fabbri, F.; Amadori, D.; De Giorgi, U. Chromogranin A predicts outcome in prostate cancer patients treated with abiraterone. Endocr. Relat. Cancer 2014, 21, 487–493. [Google Scholar] [CrossRef]
- Conteduca, V.; Aieta, M.; Amadori, D.; De Giorgi, U. Neuroendocrine differentiation in prostate cancer: Current and emerging therapy strategies. Crit. Rev. Oncol. Hematol. 2014, 92, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Conteduca, V.; Oromendia, C.; Eng, K.W.; Bareja, R.; Sigouros, M.; Molina, A.; Faltas, B.M.; Sboner, A.; Mosquera, J.M.; Elemento, O.; et al. Clinical features of neuroendocrine prostate cancer. Eur. J. Cancer 2019, 121, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Conteduca, V.; Scarpi, E.; Salvi, S.; Casadio, V.; Lolli, C.; Gurioli, G.; Schepisi, G.; Wetterskog, D.; Farolfi, A.; Menna, C.; et al. Plasma androgen receptor and serum chromogranin A in advanced prostate cancer. Sci. Rep. 2018, 8, 15442. [Google Scholar] [CrossRef] [PubMed]
- Thadani-Mulero, M.; Nanus, D.M.; Giannakakou, P. Androgen receptor on the move: Boarding the microtubule expressway to the nucleus. Cancer Res. 2012, 72, 4611–4615. [Google Scholar] [CrossRef]
- Conteduca, V.; Jayaram, A.; Romero-Laorden, N.; Wetterskog, D.; Salvi, S.; Gurioli, G.; Scarpi, E.; Castro, E.; Marin-Aguilera, M.; Lolli, C.; et al. Plasma androgen receptor and docetaxel for metastatic castration-resistant prostate cancer. Eur. Urol. 2019, 75, 368–373. [Google Scholar] [CrossRef]
- Conteduca, V.; Castro, E.; Wetterskog, D.; Scarpi, E.; Jayaram, A.; Romero-Laorden, N.; Olmos, D.; Gurioli, G.; Lolli, C.; Sáez, M.I.; et al. Plasma AR status and cabazitaxel in heavily treated metastatic castration-resistant prostate cancer. Eur. J. Cancer 2019, 116, 158–168. [Google Scholar] [CrossRef]
- De Giorgi, U.; Severi, S.; Sarnelli, A.; Sansovini, M.; Monti, M.; Gurioli, G.; Nicolini, S.; Scarpi, E.; Casadei, C.; Conteduca, V.; et al. Circulating androgen receptor (AR) gene amplification and resistance to 177Lu-PSMA-617 in patients (pts) with metastatic castration-resistant prostate cancer (mCRPC): Results of a phase II clinical trial. J. Clin. Oncol 2019, 3. [Google Scholar] [CrossRef]
- Clarke, N.; Wiechno, P.; Alekseev, B.; Sala, N.; Jones, R.; Kocak, I.; Chiuri, V.E.; Jassem, J.; Fléchon, A.; Redfern, C.; et al. Olaparib combined with abiraterone in patients with metastatic castration-resistant prostate cancer: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 2018, 19, 975–986. [Google Scholar] [CrossRef]
- Rathkopf, D.E.; Smith, M.R.; Ryan, C.J.; Berry, W.R.; Shore, N.D.; Liu, G.; Higano, C.S.; Alumkal, J.J.; Hauke, R.; Tutrone, R.F.; et al. Androgen receptor mutations in patients with castration-resistant prostate cancer treated with apalutamide. Ann. Oncol. 2017, 28, 2264–2271. [Google Scholar] [CrossRef]
- Borgmann, H.; Lallous, N.; Ozistanbullu, D.; Beraldi, E.; Paul, N.; Dalal, K.; Fazli, L.; Haferkamp, A.; Lejeune, P.; Cherkasov, A.; et al. Moving towards precision urologic oncology: Targeting enzalutamide-resistant prostate cancer and mutated forms of the androgen receptor using the novel inhibitor darolutamide (ODM-201). Eur. Urol. 2018, 73, 4–8. [Google Scholar] [CrossRef]
- Sugawara, T.; Baumgart, S.J.; Nevedomskaya, E.; Reichert, K.; Steuber, H.; Lejeune, P.; Mumberg, D.; Haendler, B. Darolutamide is a potent androgen receptor antagonist with strong efficacy in prostate cancer models. Int. J. Cancer 2019, 145, 1382–1394. [Google Scholar] [CrossRef] [PubMed]
- Conteduca, V.; Scarpi, E.; Matteucci, F.; Caroli, P.; Ravaglia, G.; Fantini, L.; Gurioli, G.; Schepisi, G.; Wetterskog, D.; Menna, C.; et al. Multimodal approach to outcome prediction in metastatic castration-resistant prostate cancer by integrating functional imaging and plasma DNA analysis. JCO Precis. Oncol. 2019, 3, 1–13. [Google Scholar] [CrossRef]


| Method | Advantages | Limitations |
|---|---|---|
| Focused | ||
| Quantitative real time PCR | Variable sensitivity with detection limit <1% (0.01% for digital PCR, PAP-A and BEAMing) Easy and rapid to use. Less expensive. Multiplex ddPCR can simultaneous screening for multiple mutations from the same sample | Necessary known hotspots in selected genes (or single probes for rare variants designed on a ‘personalized’ basis). |
| Fluorescence- labeled PCR | ||
| Nested real time PCR | ||
| ARMS-Scorpion PCR | ||
| PAP-A | ||
| BEAMing | ||
| ddPCR | ||
| Microfluidic digital PCR | ||
| Mass spectrometry | ||
| Targeted | ||
| PARE | High depth and sensitivity of analysis with detection limit 2% (0.1% and 0.01% for Tam-Seq and CAPP-Seq, respectively). De novo mutation identification. More comprehensive analysis across wider genomic regions | Very costly Requirement for high-quality DNA Extensive data analysis requiring a dedicated bioinformatician |
| Tam-Seq | ||
| Safe-Seq | ||
| CAPP-Seq | ||
| Ion-Ampliseq | ||
| Broad | ||
| WES/WGS | High sensitivity with detection limits 1–5%. Characterization a large spectrum of the genome, without the need to focus on predefined or existing alterations. More in-depth interrogation of multiple regions (WES > WGS) | Very costly Requirement for high-quality DNA Extensive data analysis requiring a dedicated bioinformatician. To accurately detect clinical mutations, a 100-to 200-fold sequencing coverage (number of times the genome is sequenced) may be needed, which are both time and cost prohibitive. Low sensitivity for the identification of copy number variation (WES) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conteduca, V.; Gurioli, G.; Brighi, N.; Lolli, C.; Schepisi, G.; Casadei, C.; Burgio, S.L.; Gargiulo, S.; Ravaglia, G.; Rossi, L.; et al. Plasma Androgen Receptor in Prostate Cancer. Cancers 2019, 11, 1719. https://doi.org/10.3390/cancers11111719
Conteduca V, Gurioli G, Brighi N, Lolli C, Schepisi G, Casadei C, Burgio SL, Gargiulo S, Ravaglia G, Rossi L, et al. Plasma Androgen Receptor in Prostate Cancer. Cancers. 2019; 11(11):1719. https://doi.org/10.3390/cancers11111719
Chicago/Turabian StyleConteduca, Vincenza, Giorgia Gurioli, Nicole Brighi, Cristian Lolli, Giuseppe Schepisi, Chiara Casadei, Salvatore Luca Burgio, Stefania Gargiulo, Giorgia Ravaglia, Lorena Rossi, and et al. 2019. "Plasma Androgen Receptor in Prostate Cancer" Cancers 11, no. 11: 1719. https://doi.org/10.3390/cancers11111719