Targeted UPLC-MS Metabolic Analysis of Human Faeces Reveals Novel Low-Invasive Candidate Markers for Colorectal Cancer
Abstract
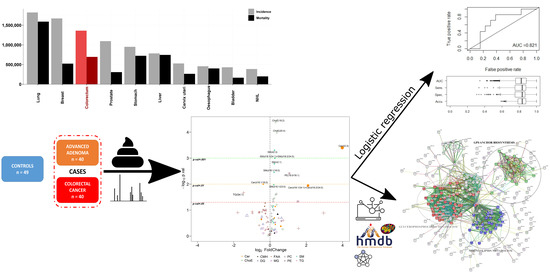
1. Introduction
2. Results
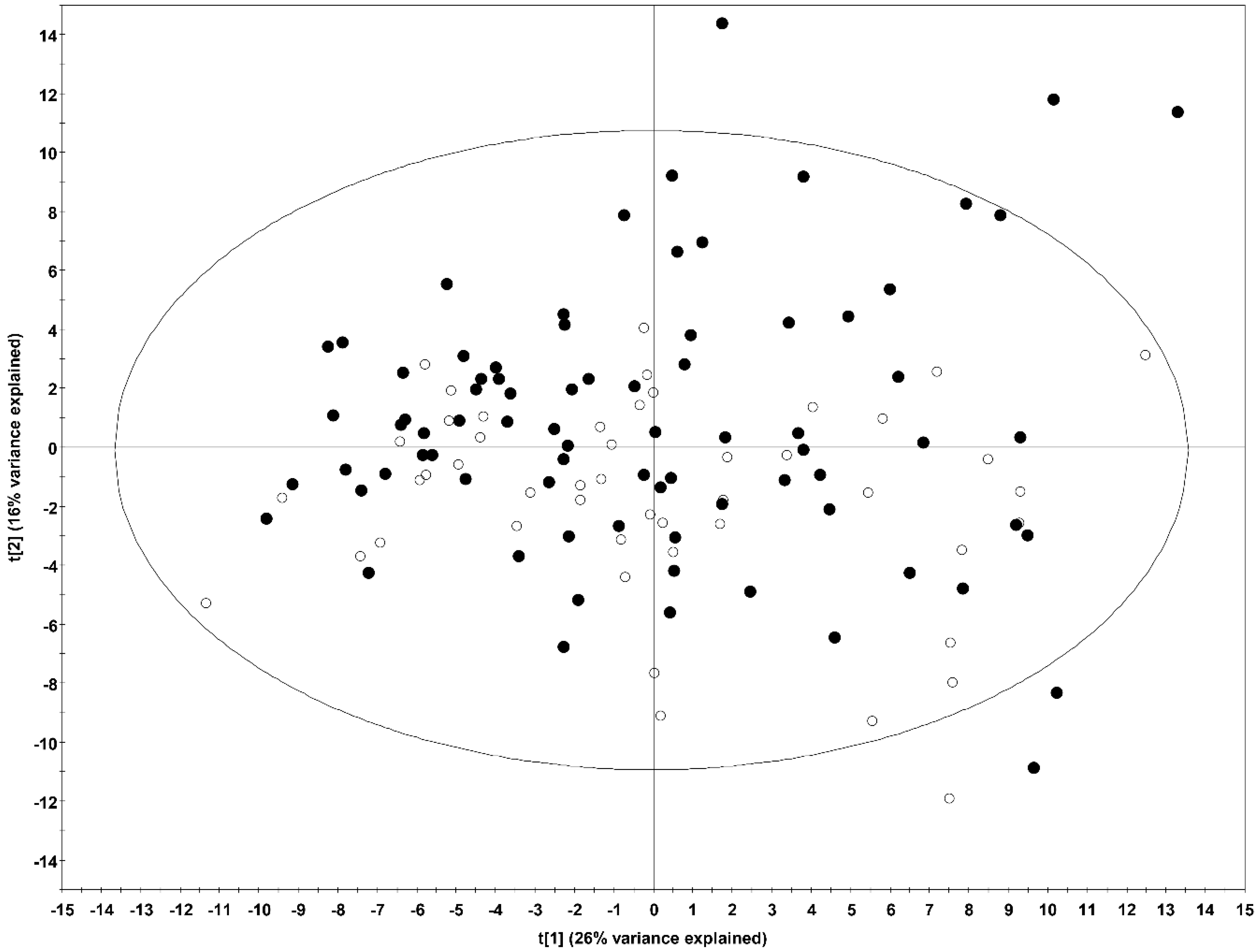
2.1. Multivariate Analysis
2.2. Univariate Analysis
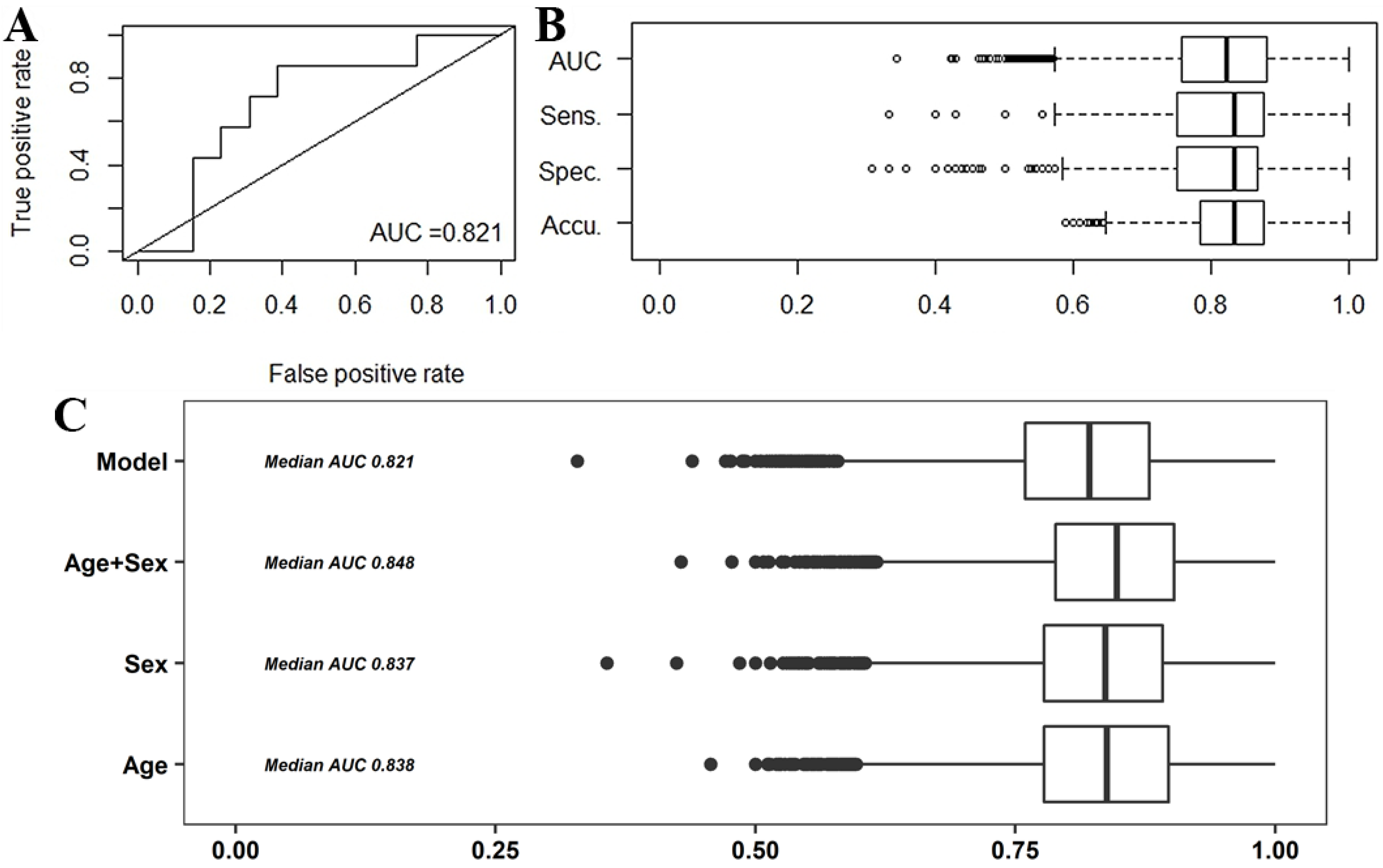
2.2.1. Predictive Models
2.2.2. Correlation of the Metabolites with Clinical Parameters
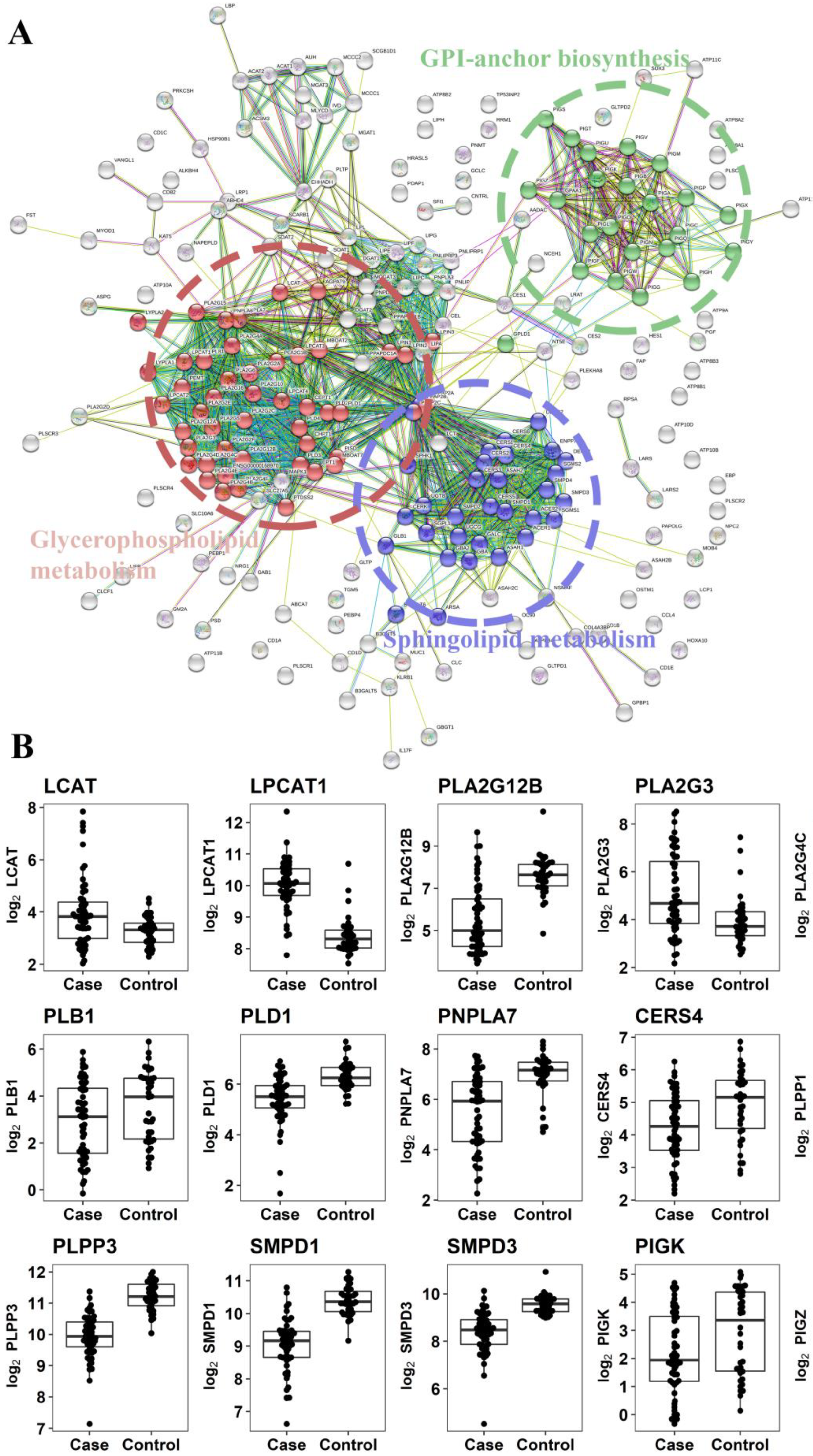
2.2.3. Gene Expression Analysis of Enzymes Involved in the Metabolism of Altered Metabolites
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Clinical Samples and Study Population
4.3. Sample Preparation and UPLC®-MS Metabolomics Analysis
4.4. Data Pre-Processing
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AUC | Area Under the Curve |
| CEA | Carcinoembryonic Antigen |
| Cer | Ceramides |
| ChoE | Cholesteryl esters |
| CMH | Monohexosylceramides |
| DAG | Diacylglycerides |
| DAPC | Diacylglycerophosphocholines |
| FAA | Fatty acid amides (Primary Fatty Amides) |
| FOB | Faecal Occult Blood |
| MAG | Monoacylglycerides |
| MEMAPC | 1-ether, 2-acylglycerophosphocholines |
| PC | Phosphatidylcholines |
| PCA | Principal Component Analysis |
| PE | Phosphatidylethanolamines |
| PI | Phosphatidylinositols |
| PLS-DA | Partial Least Square Discriminant Analysis |
| SM | Sphingomyelins |
| TAG | Triacylglycerides |
| UPLC®-MS | Ultra performance liquid chromatography-mass spectrometry |
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer Genome Landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Zauber, A.G.; Winawer, S.J.; O’Brien, M.J.; Lansdrop-Vogelaar, I.; van Ballegooijen, M.; Hankey, B.F.; Shi, W.; Bond, J.H.; Schapiro, M.; Panish, J.F.; et al. Colonoscopic Polypectomy and Long-Term Prevention of Colorectal-Cancer Deaths. N. Engl. J. Med. 2012, 366. [Google Scholar] [CrossRef] [PubMed]
- Quintero, E.; Castells, A.; Bujanda, L.; Cubiella, J.; Salas, D.; Lanas, Á.; Andreu, M.; Hernández, C.; Jover, R.; Montalvo, I.; et al. Colonoscopy versus Fecal Immunochemical Testing in Colorectal-Cancer Screening. N. Engl. J. Med. 2015, 366, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Lindholm, E.; Brevinge, H.; Haglind, E. Survival benefit in a randomized clinical trial of faecal occult blood screening for colorectal cancer. Br. J. Surg. 2008, 95, 1029–1036. [Google Scholar] [CrossRef] [PubMed]
- Faivre, J.; Dancourt, V.; Lejeune, C.; Tazi, M.A.; Lamour, J.; Gerard, D.; Dassonville, F.; Bonithon-Kopp, C. Reduction in colorectal cancer mortality by fecal occult blood screening in a French controlled study. Gastroenterology 2004, 126, 1674–1680. [Google Scholar] [CrossRef] [PubMed]
- Atkin, W.S.; Edwards, R.; Kralj-Hans, I.; Wooldrage, K.; Hart, A.R.; Northover, J.M.; Parkin, D.M.; Wardle, J.; Duffy, S.W.; Cuzick, J. Once-only flexible sigmoidoscopy screening in prevention of colorectal cancer: A multicentre randomised controlled trial. Lancet 2010, 375, 1624–1633. [Google Scholar] [CrossRef]
- Segnan, N.; Senore, C.; Andreoni, B.; Arrigoni, A.; Bisanti, L.; Cardelli, A.; Castiglione, G.; Crosta, C.; DiPlacido, R.; Ferrari, A.; et al. Randomized trial of different screening strategies for colorectal cancer: Patient response and detection rates. J. Natl. Cancer Inst. 2005, 97, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Imperiale, T.F.; Ransohoff, D.F.; Itzkowitz, S.H.; Levin, T.R.; Lavin, P.; Lidgard, G.P.; Ahlquist, D.A.; Berger, B.M. Multitarget Stool DNA Testing for Colorectal-Cancer Screening. N. Engl. J. Med. 2014, 370, 1287–1297. [Google Scholar] [CrossRef] [PubMed]
- Levin, B.; Lieberman, D.A.; McFarland, B.; Smith, R.A.; Brooks, D.; Andrews, K.S.; Dash, C.; Giardiello, F.M.; Glick, S.; Levin, T.R.; et al. Screening and Surveillance for the Early Detection of Colorectal Cancer and Adenomatous Polyps, 2008: A Joint Guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. CA Cancer J. Clin. 2008, 58, 130–160. [Google Scholar] [CrossRef] [PubMed]
- Regula, J.; Rupinski, M.; Kraszewska, E.; Polkowski, M.; Pachlewski, J.; Orlowska, J.; Nowacki, M.P.; Butruk, E. Colonoscopy Screening for Detection of Advanced Neoplasia. N. Engl. J. Med. 2006, 355, 1863–1872. [Google Scholar] [CrossRef] [PubMed]
- Bujanda, L.; Sarasqueta, C.; Zubiaurre, L.; Cosme, A.; Muñoz, C.; Sánchez, A.; Martín, C.; Tito, L.; Piñol, V.; Castells, A.; et al. Low adherence to colonoscopy in the screening of first-degree relatives of patients with colorectal cancer. Gut 2007, 56, 1714–1718. [Google Scholar] [CrossRef] [PubMed]
- Puente Gutiérrez, J.J.; Marín Moreno, M.A.; Domínguez Jiménez, J.L.; Bernal Blanco, E.; Díaz Iglesias, J.M. Effectiveness of a colonoscopic screening programme in first-degree relatives of patients with colorectal cancer. Color. Dis. 2011, 13, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, D.; McMillan, D.C.; Crearie, C.; Morrison, D.S.; Crighton, E.M.; Horgan, P.G. Temporal trends in mode, site and stage of presentation with the introduction of colorectal cancer screening: A decade of experience from the West of Scotland. Br. J. Cancer 2015, 113, 556–561. [Google Scholar] [CrossRef] [PubMed]
- Cubiella, J.; Digby, J.; Rodríguez-Alonso, L.; Vega, P.; Salve, M.; Díaz-Ondina, M.; Strachan, J.A.; Mowat, C.; McDonald, P.J.; Carey, F.A.; et al. The fecal hemoglobin concentration, age and sex test score: Development and external validation of a simple prediction tool for colorectal cancer detection in symptomatic patients. Int. J. Cancer 2017, 140, 2201–2211. [Google Scholar] [CrossRef] [PubMed]
- Cubiella, J.; Vega, P.; Salve, M.; Díaz-Ondina, M.; Alves, M.T.; Quintero, E.; Álvarez-Sánchez, V.; Fernández-Bañares, F.; Boadas, J.; Campo, R.; et al. COLONPREDICT study investigators Development and external validation of a faecal immunochemical test-based prediction model for colorectal cancer detection in symptomatic patients. BMC Med. 2016, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Westwood, M.; Lang, S.; Armstrong, N.; van Turenhout, S.; Cubiella, J.; Stirk, L.; Ramos, I.C.; Luyendijk, M.; Zaim, R.; Kleijnen, J.; et al. Faecal immunochemical tests (FIT) can help to rule out colorectal cancer in patients presenting in primary care with lower abdominal symptoms: A systematic review conducted to inform new NICE DG30 diagnostic guidance. BMC Med. 2017, 15, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Gonzalez, F.J.; Idle, J.R. LC-MS-based metabolomics in drug metabolism. Drug Metab. Rev. 2007, 39, 581–597. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.J.; Haselden, J.N. Metabolic Profiling as a Tool for Understanding Mechanisms of Toxicity. Toxicol. Pathol. 2008, 36, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Trethewey, R.N.; Krotzky, A.J.; Willmitzer, L. Metabolite profiling: from diganostics to systems biology. Nat. Rev. Mol. Cell Biol. 2004, 5, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Wilson, I.D. Understanding “global” systems biology: Metabonomics and the continuum of metabolism. Nat. Rev. Drug Discov. 2003, 2, 668–676. [Google Scholar] [CrossRef] [PubMed]
- Nordström, A.; O’Maille, G.; Qin, C.; Siuzdak, G. Non-linear Data Alignment for UPLC-MS and HPLC-MS based Metabolomics: Application to Endogenous and Exogenous Metabolites in Human Serum. Anal Chem 2006, 15, 3289–3295. [Google Scholar] [CrossRef] [PubMed]
- Nováková, L.; Solichová, D.; Solich, P. Advantages of ultra performance liquid chromatography over high-performance liquid chromatography: Comparison of different analytical approaches during analysis of diclofenac gel. J. Sep. Sci. 2006, 29, 2433–2443. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhang, Y.; Zhao, W.; Deng, K.; Wang, Z.; Yang, C.; Ma, L.; Openkova, M.S.; Hou, Y.; Li, K. Metabolomics for biomarker discovery in the diagnosis, prognosis, survival and recurrence of colorectal cancer: a systematic review. Oncotarget 2017, 8, 35460–35472. [Google Scholar] [CrossRef] [PubMed]
- Cross, A.J.; Moore, S.C.; Boca, S.; Huang, W.-Y.; Xiong, X.; Stolzenberg-Solomon, R.; Sinha, R.; Sampson, J.N. A prospective study of serum metabolites and colorectal cancer risk. Cancer 2014, 120, 3049–3057. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, A.; Nishiumi, S.; Shinohara, M.; Yoshie, T.; Hatano, N.; Okuno, T.; Bamba, T.; Fukusaki, E.; Takenawa, T.; Azuma, T.; et al. Serum metabolomics as a novel diagnostic approach for gastrointestinal cancer. Biomed. Chromatogr. 2012, 26, 548–558. [Google Scholar] [CrossRef] [PubMed]
- Leichtle, A.B.; Nuoffer, J.M.; Ceglarek, U.; Kase, J.; Conrad, T.; Witzigmann, H.; Thiery, J.; Fiedler, G.M. Serum amino acid profiles and their alterations in colorectal cancer. Metabolomics 2012, 8, 643–653. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Qin, X.; Chen, H.; Qiu, L.; Guo, Y.; Liu, H.; Chen, G.; Song, G.; Wang, X.; Li, F.; et al. Lipid profiling for early diagnosis and progression of colorectal cancer using direct-infusion electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun. Mass Spectrom. 2013, 27, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Nishiumi, S.; Kobayashi, T.; Ikeda, A.; Yoshie, T.; Kibi, M.; Izumi, Y.; Okuno, T.; Hayashi, N.; Kawano, S.; Takenawa, T.; et al. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS ONE 2012, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, P.; Wang, F.; Liu, W.; Yang, J.; Qin, H. An integrated proteomics and metabolomics approach for defining oncofetal biomarkers in the colorectal cancer. Ann. Surg. 2012, 255, 720–730. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, S.A.; Ahiahonu, P.W.K.; Jayasinghe, D.; Heath, D.; Liu, J.; Lu, Y.; Jin, W.; Kavianpour, A.; Yamazaki, Y.; Khan, A.M.; et al. Reduced levels of hydroxylated, polyunsaturated ultra long-chain fatty acids in the serum of colorectal cancer patients: implications for early screening and detection. BMC Med. 2010, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Qiu, Y.; Zou, X.; Chen, T.; Xie, G.; Cheng, Y.; Dong, T.; Zhao, L.; Feng, B.; Hu, X.; et al. Metabonomics Identi fi es Serum Metabolite Markers of Colorectal Cancer. J. Proteome Res. 2013, 12, 3000–3009. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Djukovic, D.; Deng, L.; Gu, H.; Himmati, F.; Chiorean, E.G.; Raftery, D. Colorectal cancer detection using targeted serum metabolic profiling. J. Proteome Res. 2014, 13, 4120–4130. [Google Scholar] [CrossRef] [PubMed]
- Manna, S.K.; Tanaka, N.; Krausz, K.W.; Haznadar, M.; Xue, X.; Matsubara, T.; Bowman, E.D.; Fearon, E.R.; Harris, C.C.; Shah, Y.M.; et al. Biomarkers of coordinate metabolic reprogramming in colorectal tumors in mice and humans. Gastroenterology 2014, 146, 1313–1324. [Google Scholar] [CrossRef] [PubMed]
- Mirnezami, R.; Jiménez, B.; Li, J.V.; Kinross, J.M.; Veselkov, K.; Goldin, R.D.; Holmes, E.; Nicholson, J.K.; Darzi, A. Rapid diagnosis and staging of colorectal cancer via high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy of intact tissue biopsies. Ann. Surg. 2014, 259, 1138–1149. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, L.; Zhang, H.; Deng, P.; Chen, J.; Zhou, B.; Hu, J.; Zou, J.; Lu, W.; Xiang, P.; et al. 1H NMR-based metabolic profiling of human rectal cancer tissue. Mol. Cancer 2013, 12, 121. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.L.; Passos, M.; Cmara, J.S. Investigation of urinary volatile organic metabolites as potential cancer biomarkers by solid-phase microextraction in combination with gas chromatography-mass spectrometry. Br. J. Cancer 2011, 105, 1894–1904. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Ma, C.; Liu, C.; Wang, Z.; Yang, J.; Liu, X.; Shen, Z.; Wu, R. NMR-based fecal metabolomics fingerprinting as predictors of earlier diagnosis in patients with colorectal cancer. Oncotarget 2016, 7, 29454–29464. [Google Scholar] [CrossRef] [PubMed]
- Irrazábal, T.; Belcheva, A.; Girardin, S.E.; Martin, A.; Philpott, D.J. The multifaceted role of the intestinal microbiota in colon cancer. Mol. Cell 2014, 54, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Qin, H. Microbiota disbiosis is associated with colorectal cancer. Front. Microbiol. 2015, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Li, L.; Zhu, B.; Li, Y. Lipidome in colorectal cancer. Oncotarget 2016, 7, 33429–33439. [Google Scholar] [CrossRef] [PubMed]
- Valcz, G.; Patai, Á.V.; Kalmár, A.; Péterfia, B.; Furi, I.; Wichmann, B.; Muzes, G.; Sipos, F.; Krenács, T.; Mihály, E.; et al. Myofibroblast-derived SFRP1 as potential inhibitor of colorectal carcinoma field effect. PLoS ONE 2014, 9, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.; Panasevich, M.R.; Alexander, D.; Vester Boler, B.M.; Rossoni Serao, M.C.; Faber, T.A.; Bauer, L.L.; Fahey, G.C. Fecal metabolomics of healthy breast-fed versus formula-fed infants before and during in vitro batch culture fermentation. J. Proteome Res. 2014, 13, 2534–2542. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Xie, G.; Zhao, A.; Zhao, L.; Yao, C.; Chiu, N.H.L.; Zhou, Z.; Bao, Y.; Jia, W.; Nicholson, J.K.; et al. The Footprints of Gut Microbial-Mammalian Co-Metabolism. J. Proteome Res. 2011, 10, 5512–5522. [Google Scholar] [CrossRef] [PubMed]
- Jump, R.L.P.; Polinkovsky, A.; Hurless, K.; Sitzlar, B.; Eckart, K.; Tomas, M.; Deshpande, A.; Nerandzic, M.M.; Donskey, C.J. Metabolomics analysis identifies intestinal microbiota-derived biomarkers of colonization resistance in clindamycin-treated mice. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.J.; Sprenger, N.; Montoliu, I.; Rezzi, S.; Kochhar, S.; Nicholson, J.K. Dietary Modulation of Gut Functional Ecology Studied by Fecal Metabonomics Francois-Pierre. J. Proteome Res. 2010, 9, 5284–5295. [Google Scholar] [CrossRef] [PubMed]
- Phua, L.C.; Koh, P.K.; Cheah, P.Y.; Ho, H.K.; Chan, E.C.Y. Global gas chromatography/time-of-flight mass spectrometry (GC/TOFMS)-based metabonomic profiling of lyophilized human feces. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2013, 937, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Saric, J.; Wang, Y.; Li, J.; Coen, M.; Utzinger, J.; Marchesi, J.R.; Keiser, J.; Veselkov, K.; Lindon, J.C.; Nicholson, J.K.; et al. Species variation in the fecal metabolome gives insight into differential gastrointestinal function. J. Proteome Res. 2008, 7, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Stella, C.; Beckwith-Hall, B.; Cloarec, O.; Holmes, E.; Lindon, J.C.; Powell, J.; Van Der Ouderaa, F.; Bingham, S.; Cross, A.J.; Nicholson, J.K. Susceptibility of human metabolic phenotypes to dietary modulation. J. Proteome Res. 2006, 5, 2780–2788. [Google Scholar] [CrossRef] [PubMed]
- Weir, T.L.; Manter, D.K.; Sheflin, A.M.; Barnett, B.A.; Heuberger, A.L.; Ryan, E.P. Stool Microbiome and Metabolome Differences between Colorectal Cancer Patients and Healthy Adults. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Chen, D.; Wang, N.; Zhang, T.; Zhou, R.; Huan, T.; Lu, Y.; Su, X.; Xie, Q.; Li, L.; et al. Development of High-Performance Chemical Isotope Labeling LC-MS for Profiling the Human Fecal Metabolome. Anal. Chem. 2017, 89, 6758–6765. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wu, J.; Li, J.V.; Zhou, N.; Tang, H.; Wang, Y. Gut Microbiota Composition Modifies Fecal Metabolic Profiles in Mice. J. Proteome Res. 2013, 12. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Pujos-Guillot, E.; Martin, J.F.; Galan, P.; Juste, C.; Jia, W.; Sebedio, J.L. Metabolite analysis of human fecal water by gas chromatography/mass spectrometry with ethyl chloroformate derivatization. Anal. Biochem. 2009, 393, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Pujos-Guillot, E.; Sébédio, J.L. Development of a quantitative metabolomic approach to study clinical human fecal water metabolome based on trimethylsilylation derivatization and GC/MS analysis. Anal. Chem. 2010, 82, 6447–6456. [Google Scholar] [CrossRef] [PubMed]
- Poroyko, V.; Morowitz, M.; Bell, T.; Ulanov, A.; Wang, M.; Donovan, S.; Bao, N.; Gu, S.; Hong, L.; Alverdy, J.C.; et al. Diet creates metabolic niches in the “inmature gut” that shape microbial communities. Nutr. Hosp. 2011, 26, 1283–1295. [Google Scholar] [CrossRef] [PubMed]
- Ponnusamy, K.; Choi, J.N.; Kim, J.; Lee, S.Y.; Lee, C.H. Microbial community and metabolomic comparison of irritable bowel syndrome faeces. J. Med. Microbiol. 2011, 60, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Sciorra, V.A.; Morris, A.J. Roles for lipid phosphate phosphatases in regulation of cellular signaling. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2002, 1582, 45–51. [Google Scholar] [CrossRef]
- Tang, X.; Benesch, M.G.K.; Brindley, D.N. Lipid phosphate phosphatases and their roles in mammalian physiology and pathology. J. Lipid Res. 2015, 56, 2048–2060. [Google Scholar] [CrossRef] [PubMed]
- Weaver, G.A.; Krause, J.A.; Miller, T.L.; Wolin, M.J. Short chain fatty acid distribution of enema samples from a sigmoidoscopy population:an association of high acetate and low butyrate ratios with adenomatous polyps and colon cancer. Gut 1988, 29, 1539–1543. [Google Scholar] [CrossRef] [PubMed]
- Barr, J.; Caballería, J.; Martínez-Arranz, I.; Domínguez-Díez, A.; Alonso, C.; Muntané, J.; Pérez-Cormenzana, M.; García-Monzón, C.; Mayo, R.; Martín-Duce, A.; et al. Obesity-dependent metabolic signatures associated with nonalcoholic fatty liver disease progression. J. Proteome Res. 2012, 11, 2521–2532. [Google Scholar] [CrossRef] [PubMed]
- Saccenti, E.; Hoefsloot, H.C.J.; Smilde, A.K.; Westerhuis, J.A.; Hendriks, M.M.W.B. Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics 2014, 10, 361–374. [Google Scholar] [CrossRef]
- Martínez-Arranz, I.; Mayo, R.; Pérez-Cormenzana, M.; Mincholé, I.; Salazar, L.; Alonso, C.; Mato, J.M. Enhancing metabolomics research through data mining. J. Proteomics 2015, 127, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef] [PubMed]
- Cokelaer, T.; Pultz, D.; Harder, L.M.; Serra-Musach, J.; Saez-Rodriguez, J.; Valencia, A. BioServices: A common Python package to access biological Web Services programmatically. Bioinformatics 2013, 29, 3241–3242. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [PubMed]
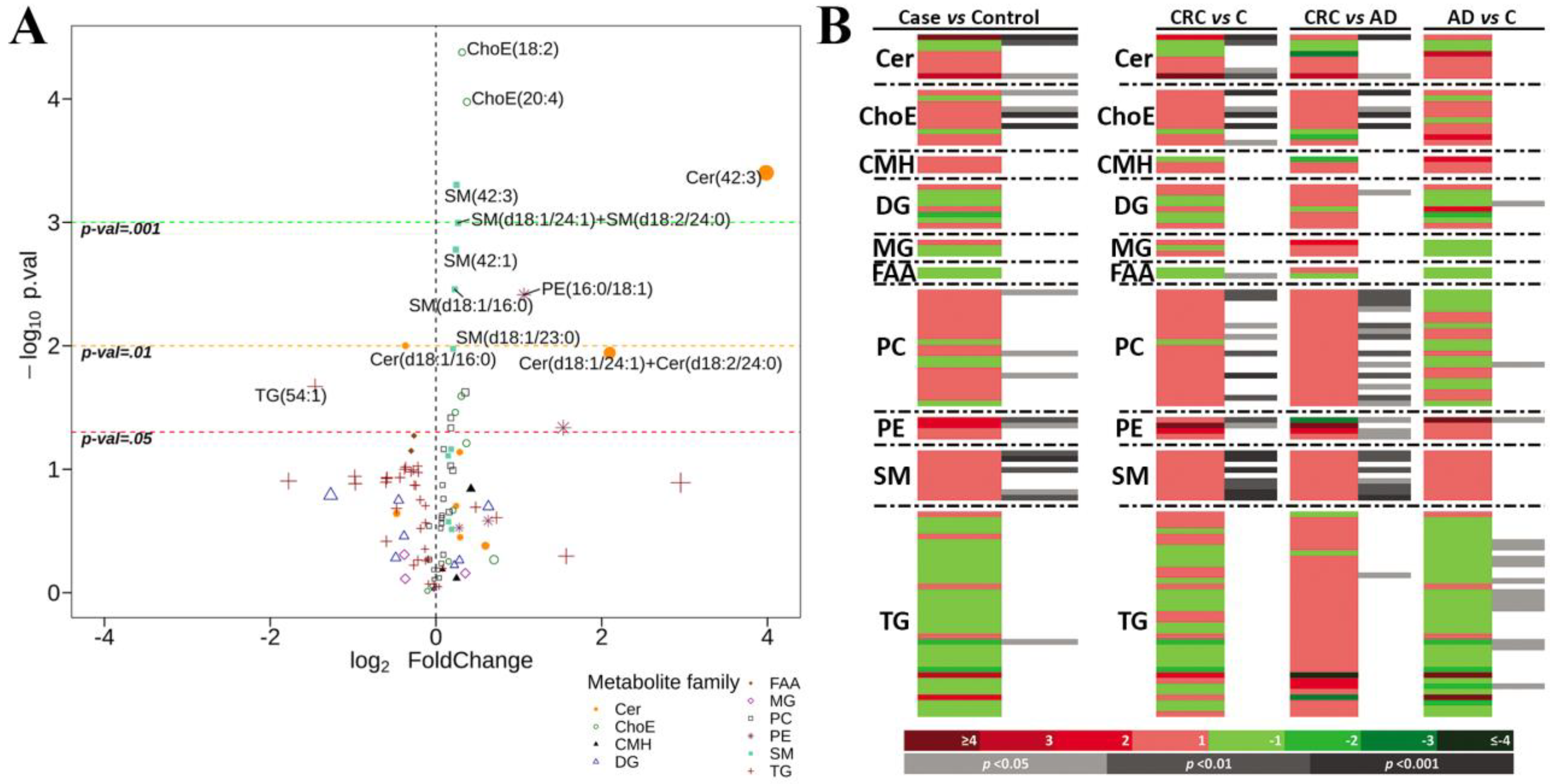
| Case vs. Control | C vs. CRC | C vs. AD | AD vs. CRC | |
|---|---|---|---|---|
| Cer (8) | 2↑ 1↓ | 3↑ 1↓ | 0 | 2↑ |
| ChoE (10) | 4↑ | 5↑ | 0 | 4↑ |
| CMH (3) | 0 | 0 | 0 | 0 |
| DG (8) | 0 | 0 | 1↓ | 1↑ |
| MG (3) | 0 | 0 | 0 | 0 |
| FAA (2) | 0 | 1↓ | 0 | 0 |
| PC (21) | 3↑ | 7↑ | 1↓ | 13↑ |
| PE (4) | 2↑ | 2↑ | 1↑ | 3↑ |
| SM (9) | 5↑ | 7↑ | 0 | 7↑ |
| TG (37) | 1↓ | 0 | 12↓ | 1↑ |
| C, AD and CRC | AvgC | AvgAD | AvgCRC | p-Value | Tukey’s HSD |
| Gender | 35.4% men | 56.4% men | 60% men | 0.042 | NA |
| Age | 62.52 | 68.64 | 73.50 | 0.0003 | CRC vs. C |
| FOB * | 0 | 49 | 873 | 1.6 × 10−9 | CRC vs. C CRC vs. AD |
| CEA | 1.90 | 1.72 | 14.85 | 0.00546 | CRC vs. C CRC vs. AD |
| COLONPREDICT | 0.048 | 0.104 | 0.470 | <2 × 10−16 | CRC vs. C CRC vs. AD |
| Control vs. Case | AvgCONTROL | AvgCASE | p-Value | ||
| Gender | 35.4% men | 58.3% men | 0.013 | ||
| Age | 62.52 | 71.10 | 0.00083 | ||
| FOB * | 0 | 336 | 7.09 × 10−8 | ||
| CEA | 1.900 | 8.367 | 0.0036 | ||
| COLONPREDICT | 0.0477 | 0.289 | 1.231 × 10−10 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cubiella, J.; Clos-Garcia, M.; Alonso, C.; Martinez-Arranz, I.; Perez-Cormenzana, M.; Barrenetxea, Z.; Berganza, J.; Rodríguez-Llopis, I.; D’Amato, M.; Bujanda, L.; et al. Targeted UPLC-MS Metabolic Analysis of Human Faeces Reveals Novel Low-Invasive Candidate Markers for Colorectal Cancer. Cancers 2018, 10, 300. https://doi.org/10.3390/cancers10090300
Cubiella J, Clos-Garcia M, Alonso C, Martinez-Arranz I, Perez-Cormenzana M, Barrenetxea Z, Berganza J, Rodríguez-Llopis I, D’Amato M, Bujanda L, et al. Targeted UPLC-MS Metabolic Analysis of Human Faeces Reveals Novel Low-Invasive Candidate Markers for Colorectal Cancer. Cancers. 2018; 10(9):300. https://doi.org/10.3390/cancers10090300
Chicago/Turabian StyleCubiella, Joaquin, Marc Clos-Garcia, Cristina Alonso, Ibon Martinez-Arranz, Miriam Perez-Cormenzana, Ziortza Barrenetxea, Jesus Berganza, Isabel Rodríguez-Llopis, Mauro D’Amato, Luis Bujanda, and et al. 2018. "Targeted UPLC-MS Metabolic Analysis of Human Faeces Reveals Novel Low-Invasive Candidate Markers for Colorectal Cancer" Cancers 10, no. 9: 300. https://doi.org/10.3390/cancers10090300
APA StyleCubiella, J., Clos-Garcia, M., Alonso, C., Martinez-Arranz, I., Perez-Cormenzana, M., Barrenetxea, Z., Berganza, J., Rodríguez-Llopis, I., D’Amato, M., Bujanda, L., Diaz-Ondina, M., & Falcón-Pérez, J. M. (2018). Targeted UPLC-MS Metabolic Analysis of Human Faeces Reveals Novel Low-Invasive Candidate Markers for Colorectal Cancer. Cancers, 10(9), 300. https://doi.org/10.3390/cancers10090300