Signal-Targeted Therapies and Resistance Mechanisms in Pancreatic Cancer: Future Developments Reside in Proteomics
Abstract
:1. Introduction
- the identification of early diagnostic and prognostic protein biomarkers;
- the identification of deregulated proteins and signalling pathways;
- the identification of protein biomarkers predictive of the response to treatment;
- the identification of early resistance mechanisms leading to the development of adapted combinatorial treatments.
1.1. Proteomics Is a Technological Tool Which Can Help to Improve the Clinical Management of Pancreatic Cancer
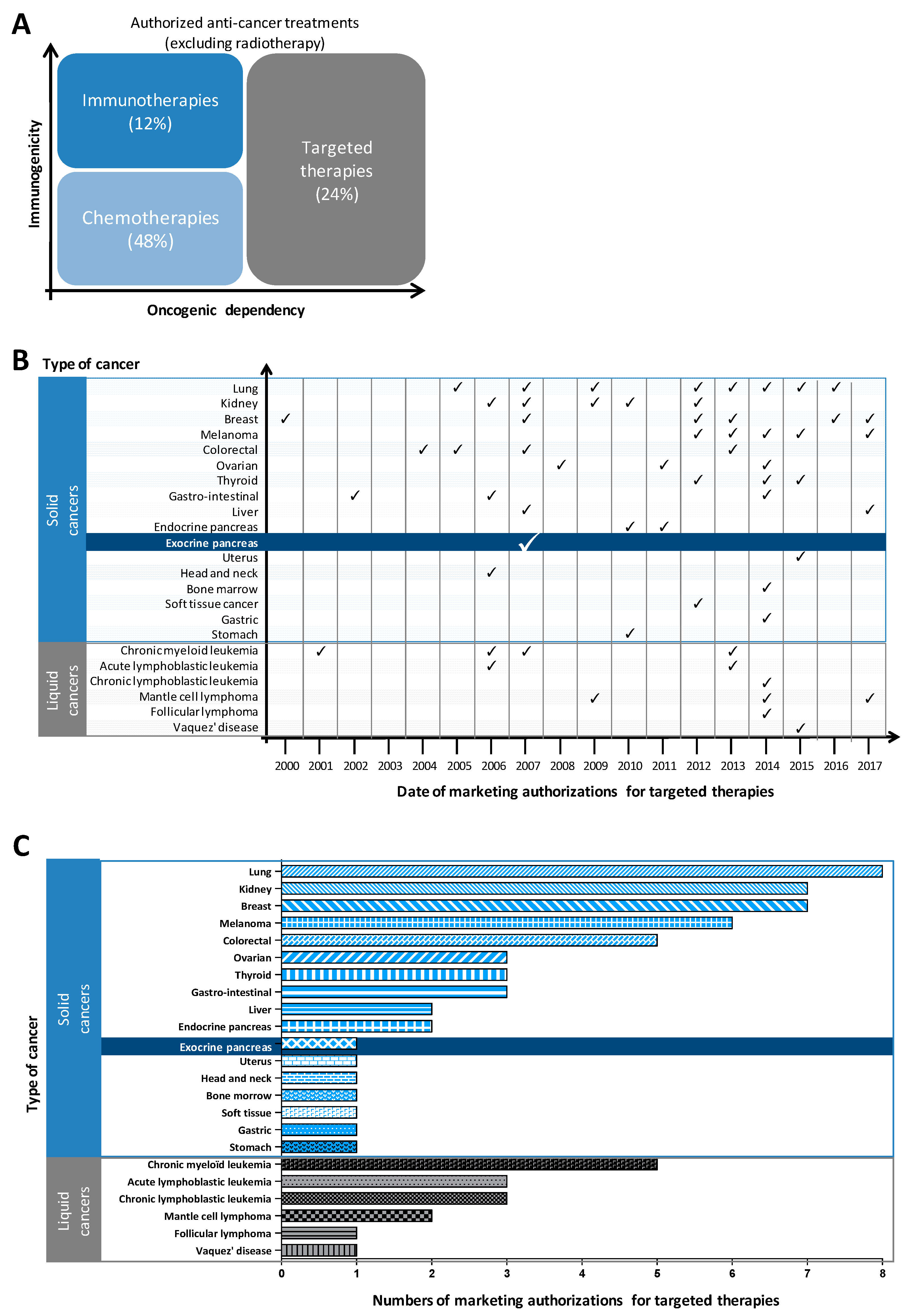
1.2. Genomic Characterization Has Failed to Identify Molecular Subtypes of Pancreatic Cancer to Guide the Choice of Targeted Therapies; Proteomics Appears to Be A Better Approach
1.3. Proteomic Profiling of PDAC Tissues
1.4. Proteomic Approaches to Search for New Biomarkers of Early Disease
1.5. Proteomic Approaches to Search for New Biomarkers of Predictive Response
1.6. Proteomic Approaches to Identify Resistance Mechanisms-Towards an Evolution in Precision Medicine
2. Conclusions/Discussion/Perspectives
- Can we use proteomics to detect PDAC earlier? Are earlier detected tumours more sensitive to targeted therapies towards PDAC oncogenic dependency (e.g., PI3K)? Will this knowledge increase life expectancy of PDAC patients?
- Can we use proteomics to refine the current (epi)-genetic and genomic characterization of PDAC to better stratify patients? Sampling patients with PDAC is difficult. Can we develop and adapt methodological work flows to aid patient sampling? These workflows should also incorporate a better understanding of the metastatic disease.
- Can we use proteomics to identify new targets (extracellular, membrane or intracellular) which take into account tumour-stroma heterotypic signalling? Is this signal different in each tumoral niche?
- Can we use proteomics to understand, at a targetable protein level/modification (phosphorylation, ubiquitination), the specific resistance of PDAC patients to targeted therapies?
Funding
Acknowledgments
Conflicts of Interest
Glossary
Abbreviations
| ALK | Anaplastic lymphoma kinase; |
| APOA2 | Apolipoprotein A2; |
| APOA4 | Apolipoprotein A4; |
| ASCO | American Society of Clinical Oncology; |
| BTK | Bruton’s tyrosine kinase; |
| CA 19-9 | Cancer antigen 19-9; |
| CDK4/6 | Cyclin-dependent kinase 4/6; |
| CSF-1R | Colony-stimulating factor 1 receptor; |
| CP | Chronic pancreatitis; |
| CTC | Circulating tumoral cell; |
| DDR1/2 | Discoidin domain receptor tyrosine kinase 1/2; |
| EGFR | Epidermal growth factor receptor; |
| ELISA | Enzyme-linked immunosorbent assay; |
| FFPE | Formalin-fixed paraffin-embedded; |
| FGFR | Fibroblast growth factor receptor; |
| FLT3 | Fms-related tyrosine kinase 3; |
| GAS6 | Growth arrest-specific 6; |
| Grb10 | Growth factor bound protein 10; |
| HER2 | human epidermal growth factor receptor 2; |
| Hsp27 | Heat shock protein 27; |
| IGF-1(R) | Insulin-like growth factor 1 (Receptor); |
| IMAC | immobilized metal affinity chromatography; |
| IPMN | Intraductal papillary mucinous neoplasm; |
| iTRAQ | isobaric tags for relative and absolute quantitation; |
| JAK | Janus kinase; |
| Lck | Lymphocyte-specific protein tyrosine kinase; |
| LCM | Laser capture microdissection; |
| MA | Marketing authorization; |
| MALDI-TOF | Matrix assisted laser desorption ionization-time of flight mass spectrometry; |
| MEK | Mitogen-activated protein kinase kinase; |
| MET | Hepatocyte growth factor receptor; |
| MRM | Multiple reaction monitoring; |
| MS | Mass spectrometry; |
| mTOR | Mammalian target of rapamycin; |
| NNRTI | Inhibitor of non-nucleoside reverse transcriptase; |
| PanIN | Pancreatic intraepithelial neoplasia; |
| PARP | Poly ADP ribose polymerase; |
| PDAC | Pancreatic ductal adenocarcinoma; |
| PDGFR | Platelet-derived growth factor receptor; |
| PDX | Patient-derived xenograft; |
| PI3K | Phosphatidylinositol-3-kinase; |
| PSCA | Prostate stem cell antigen; |
| pTyr | phosphorylated tyrosine; |
| OS | Overall survival; |
| PFS | Progression-free survival; |
| RANKL | Receptor activator of nuclear factor kappa-Β ligand; |
| ROS1 | C-ros oncogene 1; |
| RPPA | Reverse phase protein array; |
| RTK | Receptor tyrosine kinase; |
| Tie2 | Tyrosine kinase with immunoglobulin and EGF homology domains; |
| TIMP | Tissue inhibitor of metalloproteinase-1; |
| TiO2 | Titanium dioxide; |
| TGFβR | Transforming growth factor β receptor; |
| TK | Tyrosine kinase; |
| SILAC | Stable isotope labelling with amino acids in cell culture; |
| SMO | Smoothened kinase; |
| SRM | Selected reaction monitoring |
| VEGF(R) | Vascular endothelial growth factor (receptor); |
References
- Neoptolemos, J.P.; Kleeff, J.; Michl, P.; Costello, E.; Greenhalf, W.; Palmer, D.H. Therapeutic developments in pancreatic cancer: Current and future perspectives. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 333–348. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Jiang, B.; Chen, Y.; Qi, F.Z.; Zhang, J.H.; Yuan, H. Optimal adjuvant chemotherapy for resected pancreatic adenocarcinoma: A systematic review and network meta-analysis. Oncotarget 2017, 8, 81419–81429. [Google Scholar] [CrossRef] [PubMed]
- Pons-Tostivint, E.; Thibault, B.; Guillermet-Guibert, J. Targeting PI3K Signaling in Combination Cancer Therapy. Trends Cancer 2017, 3, 454–469. [Google Scholar] [CrossRef] [PubMed]
- Witkiewicz, A.K.; McMillan, E.A.; Balaji, U.; Baek, G.; Lin, W.C.; Mansour, J.; Mollaee, M.; Wagner, K.U.; Koduru, P.; Yopp, A.; et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 2015, 6, 6744. [Google Scholar] [CrossRef] [PubMed]
- Baer, R.; Cintas, C.; Dufresne, M.; Cassant-Sourdy, S.; Schonhuber, N.; Planque, L.; Lulka, H.; Couderc, B.; Bousquet, C.; Garmy-Susini, B.; et al. Pancreatic cell plasticity and cancer initiation induced by oncogenic Kras is completely dependent on wild-type PI 3-kinase p110alpha. Genes Dev. 2014, 28, 2621–2635. [Google Scholar] [CrossRef] [PubMed]
- Baer, R.; Cintas, C.; Therville, N.; Guillermet-Guibert, J. Implication of PI3K/Akt pathway in pancreatic cancer: When PI3K isoforms matter? Adv. Biol. Regul. 2015, 59, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Eser, S.; Reiff, N.; Messer, M.; Seidler, B.; Gottschalk, K.; Dobler, M.; Hieber, M.; Arbeiter, A.; Klein, S.; Kong, B.; et al. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell 2013, 23, 406–420. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.Y.; Carpenter, E.S.; Takeuchi, K.K.; Halbrook, C.J.; Peverley, L.V.; Bien, H.; Hall, J.C.; DelGiorno, K.E.; Pal, D.; Song, Y.; et al. PI3K regulation of RAC1 is required for KRAS-induced pancreatic tumorigenesis in mice. Gastroenterology 2014, 147. [Google Scholar] [CrossRef] [PubMed]
- Burris, H.A., III; Moore, M.J.; Andersen, J.; Green, M.R.; Rothenberg, M.L.; Modiano, M.R.; Cripps, M.C.; Portenoy, R.K.; Storniolo, A.M.; Tarassoff, P.; Nelson, R.; et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: A randomized trial. J. Clin. Oncol. 1997, 15, 2403–2413. [Google Scholar] [CrossRef] [PubMed]
- Adamska, A.; Domenichini, A.; Falasca, M. Pancreatic Ductal Adenocarcinoma: Current and Evolving Therapies. Int. J. Mol. Sci. 2017, 18, 1338. [Google Scholar] [CrossRef] [PubMed]
- Danovi, S.A.; Wong, H.H.; Lemoine, N.R. Targeted therapies for pancreatic cancer. Br. Med. Bull. 2008, 87, 97–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barati Bagherabad, M.; Afzaljavan, F.; ShahidSales, S.; Hassanian, S.M.; Avan, A. Targeted Therapies in Pancreatic Cancer: Promises and Failures. J. Cell. Biochem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Paulson, A.S.; Cao, H.S.T.; Tempero, M.A.; Lowy, A.M. Therapeutic advances in pancreatic cancer. Gastroenterology 2013, 144, 1316–1326. [Google Scholar] [CrossRef] [PubMed]
- Von Hoff, D.D.; Ramanathan, R.K.; Borad, M.J.; Laheru, D.A.; Smith, L.S.; Wood, T.E.; Korn, R.L.; Desai, N.; Trieu, V.; Iglesias, J.L.; et al. Gemcitabine plus nab-paclitaxel is an active regimen in patients with advanced pancreatic cancer: A phase I/II trial. J. Clin. Oncol. 2011, 29, 4548–4554. [Google Scholar] [CrossRef] [PubMed]
- De Vita, F.; Ventriglia, J.; Febbraro, A.; Laterza, M.M.; Fabozzi, A.; Savastano, B.; Petrillo, A.; Diana, A.; Giordano, G.; Troiani, T.; et al. NAB-paclitaxel and gemcitabine in metastatic pancreatic ductal adenocarcinoma (PDAC): From clinical trials to clinical practice. BMC Cancer 2016, 16, 709. [Google Scholar] [CrossRef] [PubMed]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouche, O.; Guimbaud, R.; Becouarn, Y.; Adenis, A.; Raoul, J.L.; Gourgou-Bourgade, S.; de la Fouchardiere, C.; et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef] [PubMed]
- Gunturu, K.S.; Yao, X.; Cong, X.; Thumar, J.R.; Hochster, H.S.; Stein, S.M.; Lacy, J. FOLFIRINOX for locally advanced and metastatic pancreatic cancer: Single institution retrospective review of efficacy and toxicity. Med. Oncol. 2013, 30, 361. [Google Scholar] [CrossRef] [PubMed]
- Humphery-Smith, I. A human proteome project with a beginning and an end. Proteomics 2004, 4, 2519–2521. [Google Scholar] [CrossRef] [PubMed]
- Fenn, J.B.; Mann, M.; Meng, C.K.; Wong, S.F.; Whitehouse, C.M. Electrospray ionization for mass spectrometry of large biomolecules. Science 1989, 246, 64–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bantscheff, M.; Lemeer, S.; Savitski, M.M.; Kuster, B. Quantitative mass spectrometry in proteomics: Critical review update from 2007 to the present. Anal. Bioanal. Chem. 2012, 404, 939–965. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Franc, V.; Heck, A.J.R. Glycoproteomics: A Balance between High-Throughput and In-Depth Analysis. Trends Biotechnol. 2017, 35, 598–609. [Google Scholar] [CrossRef] [PubMed]
- Heap, R.E.; Gant, M.S.; Lamoliatte, F.; Peltier, J.; Trost, M. Mass spectrometry techniques for studying the ubiquitin system. Biochem. Soc. Trans. 2017, 45, 1137–1148. [Google Scholar] [CrossRef] [PubMed]
- Hogrebe, A.; von Stechow, L.; Bekker-Jensen, D.B.; Weinert, B.T.; Kelstrup, C.D.; Olsen, J.V. Benchmarking common quantification strategies for large-scale phosphoproteomics. Nat. Commun. 2018, 9, 1045. [Google Scholar] [CrossRef] [PubMed]
- Mueller, M.; Martens, L.; Apweiler, R. Annotating the human proteome: Beyond establishing a parts list. Biochim. Biophys. Acta 2007, 1774, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Castillo, J.; Bernard, V.; Lucas, F.A.S.; Allenson, K.; Capello, M.; Kim, D.U.; Gascoyne, P.; Mulu, F.C.; Stephens, B.M.; Huang, J.; et al. Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients. Ann. Oncol. 2018, 29, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Capello, M.; Bantis, L.E.; Scelo, G.; Zhao, Y.; Li, P.; Dhillon, D.S.; Patel, N.J.; Kundnani, D.L.; Wang, H.; Abbruzzese, J.L.; et al. Sequential Validation of Blood-Based Protein Biomarker Candidates for Early-Stage Pancreatic Cancer. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef] [PubMed]
- Grote, T.; Siwak, D.R.; Fritsche, H.A.; Joy, C.; Mills, G.B.; Simeone, D.; Whitcomb, D.C.; Logsdon, C.D. Validation of reverse phase protein array for practical screening of potential biomarkers in serum and plasma: Accurate detection of CA19-9 levels in pancreatic cancer. Proteomics 2008, 8, 3051–3060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honda, K.; Kobayashi, M.; Okusaka, T.; Rinaudo, J.A.; Huang, Y.; Marsh, T.; Sanada, M.; Sasajima, Y.; Nakamori, S.; Shimahara, M.; et al. Plasma biomarker for detection of early stage pancreatic cancer and risk factors for pancreatic malignancy using antibodies for apolipoprotein-AII isoforms. Sci. Rep. 2015, 5, 15921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilies, M.; Sappa, P.K.; Iuga, C.A.; Loghin, F.; Salazar, M.G.; Weiss, F.U.; Beyer, G.; Lerch, M.M.; Volker, U.; Mayerle, J.; et al. Plasma protein profiling of patients with intraductal papillary mucinous neoplasm of the pancreas as potential precursor lesions of pancreatic cancer. Clin. Chim. Acta 2018, 477, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Ligat, L.; Saint-Laurent, N.; El-Mrani, A.; Gigoux, V.; al Saati, T.; Tomasini, R.; Nigri, J.; Dejean, S.; Pont, F.; Baer, R.; et al. Pancreatic preneoplastic lesions plasma signatures and biomarkers based on proteome profiling of mouse models. Br. J. Cancer 2015, 113, 1590–1598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crnogorac-Jurcevic, T.; Gangeswaran, R.; Bhakta, V.; Capurso, G.; Lattimore, S.; Akada, M.; Sunamura, M.; Prime, W.; Campbell, F.; Brentnall, T.A.; et al. Proteomic analysis of chronic pancreatitis and pancreatic adenocarcinoma. Gastroenterology 2005, 129, 1454–1463. [Google Scholar] [CrossRef] [PubMed]
- Fakelman, F.; Felix, K.; Buchler, M.W.; Werner, J. New pre-analytical approach for the deep proteome analysis of sera from pancreatitis and pancreas cancer patients. Arch. Physiol. Biochem. 2010, 116, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Gruner, B.M.; Hahne, H.; Mazur, P.K.; Trajkovic-Arsic, M.; Maier, S.; Esposito, I.; Kalideris, E.; Michalski, C.W.; Kleeff, J.; Rauser, S.; et al. MALDI imaging mass spectrometry for in situ proteomic analysis of preneoplastic lesions in pancreatic cancer. PLoS ONE 2012, 7, e39424. [Google Scholar] [CrossRef] [PubMed]
- Jenkinson, C.; Elliott, V.; Menon, U.; Apostolidou, S.; Fourkala, O.E.; Gentry-Maharaj, A.; Pereira, S.P.; Jacobs, I.; Cox, T.F.; Greenhalf, W.; et al. Evaluation in pre-diagnosis samples discounts ICAM-1 and TIMP-1 as biomarkers for earlier diagnosis of pancreatic cancer. J. Proteom. 2015, 113, 400–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenkinson, C.; Elliott, V.L.; Evans, A.; Oldfield, L.; Jenkins, R.E.; O’Brien, D.P.; Apostolidou, S.; Gentry-Maharaj, A.; Fourkala, E.O.; Jacobs, I.J.; et al. Decreased Serum Thrombospondin-1 Levels in Pancreatic Cancer Patients Up to 24 Months Prior to Clinical Diagnosis: Association with Diabetes Mellitus. Clin. Cancer Res. 2016, 22, 1734–1743. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Wu, W.C.; Zhao, G.C.; Wang, D.S.; Lou, W.H.; Jin, D.Y. ITRAQ-based quantitative proteomics reveals apolipoprotein A-I and transferrin as potential serum markers in CA19-9 negative pancreatic ductal adenocarcinoma. Medicine (Baltimore) 2016, 95, e4527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mustafa, S.; Pan, L.; Marzoq, A.; Fawaz, M.; Sander, L.; Ruckert, F.; Schrenk, A.; Hartl, C.; Uhler, R.; Yildirim, A.; et al. Comparison of the tumor cell secretome and patient sera for an accurate serum-based diagnosis of pancreatic ductal adenocarcinoma. Oncotarget 2017, 8, 11963–11976. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, E.; Park, K.J.; Park, H.D.; Kim, J.W.; Woo, H.I.; Lee, K.H.; Lee, K.T.; Lee, J.K.; Park, J.O.; et al. Large-scale clinical validation of biomarkers for pancreatic cancer using a mass spectrometry-based proteomics approach. Oncotarget 2017, 8, 42761–42771. [Google Scholar] [CrossRef] [PubMed]
- Saraswat, M.; Joenvaara, S.; Seppanen, H.; Mustonen, H.; Haglund, C.; Renkonen, R. Comparative proteomic profiling of the serum differentiates pancreatic cancer from chronic pancreatitis. Cancer Med. 2017, 6, 1738–1751. [Google Scholar] [CrossRef] [PubMed]
- Takano, S.; Sogawa, K.; Yoshitomi, H.; Shida, T.; Mogushi, K.; Kimura, F.; Shimizu, H.; Yoshidome, H.; Ohtsuka, M.; Kato, A.; et al. Increased circulating cell signalling phosphoproteins in sera are useful for the detection of pancreatic cancer. Br. J. Cancer 2010, 103, 223–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wehr, A.Y.; Hwang, W.T.; Blair, I.A.; Yu, K.H. Relative quantification of serum proteins from pancreatic ductal adenocarcinoma patients by stable isotope dilution liquid chromatography-mass spectrometry. J. Proteome Res. 2012, 11, 1749–1758. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Pan, S.; Cooke, K.; Moyes, K.W.; Bronner, M.P.; Goodlett, D.R.; Aebersold, R.; Brentnall, T.A. Comparison of pancreas juice proteins from cancer versus pancreatitis using quantitative proteomic analysis. Pancreas 2007, 34, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Jabbar, K.S.; Arike, L.; Verbeke, C.S.; Sadik, R.; Hansson, G.C. Highly Accurate Identification of Cystic Precursor Lesions of Pancreatic Cancer Through Targeted Mass Spectrometry: A Phase IIc Diagnostic Study. J. Clin. Oncol. 2018, 36, 367–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shekouh, A.R.; Thompson, C.C.; Prime, W.; Campbell, F.; Hamlett, J.; Herrington, C.S.; Lemoine, N.R.; Crnogorac-Jurcevic, T.; Buechler, M.W.; Friess, H.; et al. Application of laser capture microdissection combined with two-dimensional electrophoresis for the discovery of differentially regulated proteins in pancreatic ductal adenocarcinoma. Proteomics 2003, 3, 1988–2001. [Google Scholar] [CrossRef] [PubMed]
- Sitek, B.; Sipos, B.; Alkatout, I.; Poschmann, G.; Stephan, C.; Schulenborg, T.; Marcus, K.; Luttges, J.; Dittert, D.D.; Baretton, G.; et al. Analysis of the pancreatic tumor progression by a quantitative proteomic approach and immunhistochemical validation. J. Proteome Res. 2009, 8, 1647–1656. [Google Scholar] [CrossRef] [PubMed]
- Britton, D.; Zen, Y.; Quaglia, A.; Selzer, S.; Mitra, V.; Lobetaner, C.; Jung, S.; Bohm, G.; Schmid, P.; Prefot, P.; et al. Quantification of pancreatic cancer proteome and phosphorylome: Indicates molecular events likely contributing to cancer and activity of drug targets. PLoS ONE 2014, 9, e90948. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.J.; Frazier, M.L.; Zhang, N.; Liu, Q.; Wei, C. Reverse-phase protein array analysis to identify biomarker proteins in human pancreatic cancer. Dig. Dis. Sci. 2014, 59, 968–975. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.Y.; Muzumdar, M.D.; Dorans, K.J.; Robbins, R.A.; Bhutkar, A.; del Rosario, A.M.; Mertins, P.; Qiao, J.; Schafer, C.; Gertler, F.B.; et al. Adaptive and reversible resistance to Kras inhibition in pancreatic cancer cells. Cancer Res. 2018, 78, 985–1002. [Google Scholar] [CrossRef] [PubMed]
- Gruner, B.M.; Winkelmann, I.; Feuchtinger, A.; Sun, N.; Balluff, B.; Teichmann, N.; Herner, A.; Kalideris, E.; Steiger, K.; Braren, R.; et al. Modeling Therapy Response and Spatial Tissue Distribution of Erlotinib in Pancreatic Cancer. Mol. Cancer Ther. 2016, 15, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Tape, C.J.; Ling, S.; Dimitriadi, M.; McMahon, K.M.; Worboys, J.D.; Leong, H.S.; Norrie, I.C.; Miller, C.J.; Poulogiannis, G.; Lauffenburger, D.A.; et al. Oncogenic KRAS Regulates Tumor Cell Signaling via Stromal Reciprocation. Cell 2016, 165, 1818. [Google Scholar] [CrossRef] [PubMed]
- Brandi, J.; Pozza, E.D.; Dando, I.; Biondani, G.; Robotti, E.; Jenkins, R.; Elliott, V.; Park, K.; Marengo, E.; Costello, E.; et al. Secretome protein signature of human pancreatic cancer stem-like cells. J. Proteom. 2016, 136, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gronborg, M.; Kristiansen, T.Z.; Iwahori, A.; Chang, R.; Reddy, R.; Sato, N.; Molina, H.; Jensen, O.N.; Hruban, R.H.; Goggins, M.G.; et al. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol. Cell. Proteom. 2006, 5, 157–171. [Google Scholar] [CrossRef] [PubMed]
- Brandi, J.; Dando, I.; Pozza, E.D.; Biondani, G.; Jenkins, R.; Elliott, V.; Park, K.; Fanelli, G.; Zolla, L.; Costello, E.; et al. Proteomic analysis of pancreatic cancer stem cells: Functional role of fatty acid synthesis and mevalonate pathways. J. Proteom. 2017, 150, 310–322. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, E.S.; Su, S.P.; Nagrial, A.M.; Hochgrafe, F.; Pajic, M.; Lehrbach, G.M.; Parton, R.G.; Yap, A.S.; Horvath, L.G.; Chang, D.K.; et al. Resolution of Novel Pancreatic Ductal Adenocarcinoma Subtypes by Global Phosphotyrosine Profiling. Mol. Cell. Proteom. 2016, 15, 2671–2685. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Zhong, Y.; Yachida, S.; Rajeshkumar, N.V.; Abel, M.L.; Marimuthu, A.; Mudgal, K.; Hruban, R.H.; Poling, J.S.; Tyner, J.W.; et al. Heterogeneity of pancreatic cancer metastases in a single patient revealed by quantitative proteomics. Mol. Cell. Proteom. 2014, 13, 2803–2811. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Niu, J.; Li, J.; Shen, X.; Shen, S.; Straubinger, R.M.; Qu, J. Temporal Effects of Combined Birinapant and Paclitaxel on Pancreatic Cancer Cells Investigated via Large-scale, Ion-Current-Based Quantitative Proteomics (IonStar). Mol. Cell. Proteom. 2018, 17, 655–671. [Google Scholar] [CrossRef] [PubMed]
- Domon, B.; Aebersold, R. Options and considerations when selecting a quantitative proteomics strategy. Nat. Biotechnol. 2010, 28, 710–721. [Google Scholar] [CrossRef] [PubMed]
- Kosako, H.; Nagano, K. Quantitative phosphoproteomics strategies for understanding protein kinase-mediated signal transduction pathways. Expert Rev. Proteom. 2011, 8, 81–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cutillas, P.R.; Jorgensen, C. Biological signalling activity measurements using mass spectrometry. Biochem. J. 2011, 434, 189–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sikkema, A.H.; den Dunnen, W.F.; Diks, S.H.; Peppelenbosch, M.P.; de Bont, E.S. Optimizing targeted cancer therapy: Towards clinical application of systems biology approaches. Crit. Rev. Oncol. Hematol. 2012, 82, 171–186. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.J.; Goldstein, D.; Hamm, J.; Figer, A.; Hecht, J.R.; Gallinger, S.; Au, H.J.; Murawa, P.; Walde, D.; Wolff, R.A.; et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: A phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J. Clin. Oncol. 2007, 25, 1960–1966. [Google Scholar] [CrossRef] [PubMed]
- Rougier, P.; Riess, H.; Manges, R.; Karasek, P.; Humblet, Y.; Barone, C.; Santoro, A.; Assadourian, S.; Hatteville, L.; Philip, P.A. Randomised, placebo-controlled, double-blind, parallel-group phase III study evaluating aflibercept in patients receiving first-line treatment with gemcitabine for metastatic pancreatic cancer. Eur. J. Cancer 2013, 49, 2633–2642. [Google Scholar] [CrossRef] [PubMed]
- Amanam, I.; Chung, V. Targeted Therapies for Pancreatic Cancer. Cancers 2018, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Di Marco, M.; Grassi, E.; Durante, S.; Vecchiarelli, S.; Palloni, A.; Macchini, M.; Casadei, R.; Ricci, C.; Panzacchi, R.; Santini, D.; et al. State of the art biological therapies in pancreatic cancer. World J. Gastrointest. Oncol. 2016, 8, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Burrell, R.A.; Swanton, C. Tumour heterogeneity and the evolution of polyclonal drug resistance. Mol. Oncol. 2014, 8, 1095–1111. [Google Scholar] [CrossRef] [PubMed]
- Almendro, V.; Marusyk, A.; Polyak, K. Cellular heterogeneity and molecular evolution in cancer. Annu. Rev. Pathol. 2013, 8, 277–302. [Google Scholar] [CrossRef] [PubMed]
- Makohon-Moore, A.P.; Zhang, M.; Reiter, J.G.; Bozic, I.; Allen, B.; Kundu, D.; Chatterjee, K.; Wong, F.; Jiao, Y.; Kohutek, Z.A.; et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat. Genet. 2017, 49, 358–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waddell, N.; Pajic, M.; Patch, A.M.; Chang, D.K.; Kassahn, K.S.; Bailey, P.; Johns, A.L.; Miller, D.; Nones, K.; Quek, K.; et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015, 518, 495–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.M.; Gingras, M.C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Sivakumar, S.; de Santiago, I.; Chlon, L.; Markowetz, F. Master Regulators of Oncogenic KRAS Response in Pancreatic Cancer: An Integrative Network Biology Analysis. PLoS Med. 2017, 14, e1002223. [Google Scholar] [CrossRef] [PubMed]
- Collisson, E.A.; Sadanandam, A.; Olson, P.; Gibb, W.J.; Truitt, M.; Gu, S.; Cooc, J.; Weinkle, J.; Kim, G.E.; Jakkula, L.; et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011, 17, 500–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Kwok-Shing Ng, P.; Kucherlapati, M.; Chen, F.; Liu, Y.; Tsang, Y.H.; de Velasco, G.; Jeong, K.J.; Akbani, R.; Hadjipanayis, A.; et al. A Pan-Cancer Proteogenomic Atlas of PI3K/AKT/mTOR Pathway Alterations. Cancer Cell 2017, 31. [Google Scholar] [CrossRef] [PubMed]
- Cintas, C.; Guillermet-Guibert, J. Heterogeneity of Phosphatidylinositol-3-Kinase (PI3K)/AKT/Mammalian Target of Rapamycin Activation in Cancer: Is PI3K Isoform Specificity Important? Front. Oncol. 2017, 7, 330. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, X.; Lang, H.; Zhang, S.; Luo, Y.; Zhang, J. S100 calcium-binding protein A6 promotes epithelial-mesenchymal transition through beta-catenin in pancreatic cancer cell line. PLoS ONE 2015, 10, e0121319. [Google Scholar]
- Zhu, J.; He, J.; Liu, Y.; Simeone, D.M.; Lubman, D.M. Identification of glycoprotein markers for pancreatic cancer CD24+CD44+ stem-like cells using nano-LC-MS/MS and tissue microarray. J. Proteome Res. 2012, 11, 2272–2281. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Chen, Z.; He, L.; He, N.; Xi, Z.; Li, Z.; Deng, Y.; Zeng, X. Mass spectrometry-assisted gel-based proteomics in cancer biomarker discovery: Approaches and application. Theranostics 2017, 7, 3559–3572. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Park, J.; Wang, J.I.; Kim, Y. Recent advances in proteomic profiling of pancreatic ductal adenocarcinoma and the road ahead. Expert Rev. Proteom. 2017, 14, 963–971. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Brentnall, T.A.; Chen, R. Proteomics analysis of bodily fluids in pancreatic cancer. Proteomics 2015, 15, 2705–2715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wakabayashi, T.; Sawabu, N.; Takemori, Y.; Satomura, Y.; Kidani, H.; Ohta, H.; Watanabe, H.; Yamakawa, O.; Takahashi, H.; Watanabe, K.; et al. Diagnostic significance of cancer-associated carbohydrate antigen (CA19-9) concentrations in pancreatic juice: Analysis in pure pancreatic juice collected by endoscopic aspiration and immunohistochemical study in chronic pancreatitis. Pancreas 1993, 8, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Rosendahl, A.H.; Ansari, D.; Andersson, R. Proteome-based biomarkers in pancreatic cancer. World J. Gastroenterol. 2011, 17, 4845–4852. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Bamlet, W.R.; Oberg, A.L.; Chaffee, K.G.; Donahue, G.; Cao, X.J.; Chari, S.; Garcia, B.A.; Petersen, G.M.; Zaret, K.S. Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19-9 blood markers. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Nie, S.; Lo, A.; Wu, J.; Zhu, J.; Tan, Z.; Simeone, D.M.; Anderson, M.A.; Shedden, K.A.; Ruffin, M.T.; Lubman, D.M. Glycoprotein biomarker panel for pancreatic cancer discovered by quantitative proteomics analysis. J. Proteome Res. 2014, 13, 1873–1884. [Google Scholar] [CrossRef] [PubMed]
- Gillet, L.C.; Navarro, P.; Tate, S.; Rost, H.; Selevsek, N.; Reiter, L.; Bonner, R.; Aebersold, R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell. Proteom. 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Krasny, L.; Bland, P.; Kogata, N.; Wai, P.; Howard, B.A.; Natrajan, R.C.; Huang, P.H. SWATH mass spectrometry as a tool for quantitative profiling of the matrisome. J. Proteom. 2018. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.P.; Kang, S.A.; Rameseder, J.; Zhang, Y.; Ottina, K.A.; Lim, D.; Peterson, T.R.; Choi, Y.; Gray, N.S.; Yaffe, M.B.; et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 2011, 332, 1317–1322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Yoon, S.O.; Poulogiannis, G.; Yang, Q.; Ma, X.M.; Villen, J.; Kubica, N.; Hoffman, G.R.; Cantley, L.C.; Gygi, S.P.; et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 2011, 332, 1322–1326. [Google Scholar] [CrossRef] [PubMed]
- Alagesan, B.; Contino, G.; Guimaraes, A.R.; Corcoran, R.B.; Deshpande, V.; Wojtkiewicz, G.R.; Hezel, A.F.; Wong, K.K.; Loda, M.; Weissleder, R.; et al. Combined MEK and PI3K inhibition in a mouse model of pancreatic cancer. Clin. Cancer Res. 2015, 21, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Junttila, M.R.; Devasthali, V.; Cheng, J.H.; Castillo, J.; Metcalfe, C.; Clermont, A.C.; Otter, D.D.; Chan, E.; Bou-Reslan, H.; Cao, T.; et al. Modeling targeted inhibition of MEK and PI3 kinase in human pancreatic cancer. Mol. Cancer Ther. 2015, 14, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Ying, H.; Dey, P.; Yao, W.; Kimmelman, A.C.; Draetta, G.F.; Maitra, A.; DePinho, R.A. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2016, 30, 355–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalla Pozza, E.; Manfredi, M.; Brandi, J.; Buzzi, A.; Conte, E.; Pacchiana, R.; Cecconi, D.; Marengo, E.; Donadelli, M. Trichostatin A alters cytoskeleton and energy metabolism of pancreatic adenocarcinoma cells: An in depth proteomic study. J. Cell. Biochem. 2018, 119, 2696–2707. [Google Scholar] [CrossRef] [PubMed]
- Hilhorst, R.; Houkes, L.; van den Berg, A.; Ruijtenbeek, R. Peptide microarrays for detailed, high-throughput substrate identification, kinetic characterization, and inhibition studies on protein kinase A. Anal. Biochem. 2009, 387, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Shukla, H.D. Comprehensive Analysis of Cancer-Proteogenome to Identify Biomarkers for the Early Diagnosis and Prognosis of Cancer. Proteomes 2017, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Breitkreutz, D.; Hlatky, L.; Rietman, E.; Tuszynski, J.A. Molecular signaling network complexity is correlated with cancer patient survivability. Proc. Natl. Acad. Sci. USA 2012, 109, 9209–9212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bournet, B.; Vignolle-Vidoni, A.; Grand, D.; Roques, C.; Breibach, F.; Cros, J.; Muscari, F.; Carrere, N.; Selves, J.; Cordelier, P.; et al. Endoscopic ultrasound-guided fine-needle aspiration plus KRAS and GNAS mutation in malignant intraductal papillary mucinous neoplasm of the pancreas. Endosc. Int. Open 2016, 4, E1228–E1235. [Google Scholar] [CrossRef] [PubMed]

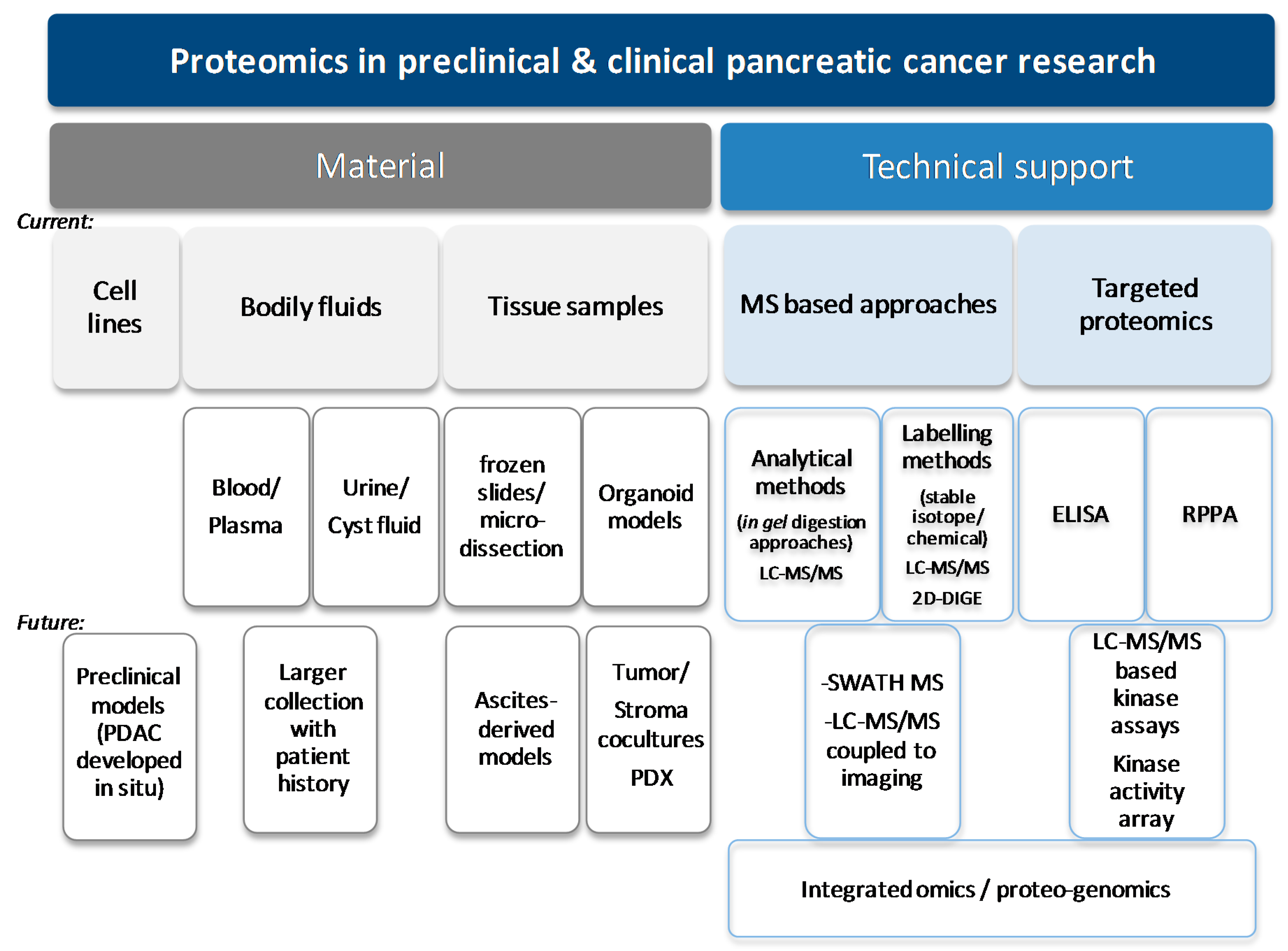
| Source | Details of Source | PDAC Clinical Application | PDAC Tumoural Biology | Actual Limitations/Developments |
|---|---|---|---|---|
| Biopsies (fine needle under echo-endoscopy) | Diagnostic Active surveillance in patients at risk (chronic pancreatitis, mucinous lesions, hereditary) |
|
| Poor cellularity Development of organoids or PDX |
| Metastasis | Loco-regional metastasis (peritoneum, ascites) Distant metastasis (lung, liver) |
|
| Limited sampling |
| Resection | Normal adjacent tissue Tumour Desmoplastic reaction |
|
| Limited to 15–20% of all PDAC patients-do not represent the most aggressive patients Development of MS coupled to imaging |
| Body fluids | Blood, blood fractions (serum, plasma, exosomes, CTCs, etc...) Urine Ascites |
|
| Selection of patients based on circulating DNA Development of ascites-based PDX as an easy access to metastatic cells in their environment (e.g., immune cells) |
| Cell lines/in situ experimental PDAC | Not applicable | Do not fully represent the heterogeneity of PDAC Study of heterotypic communication between stromal and cancer cells | ||
| Conditioned medium | Not applicable |
| Study of heterotypic communication between stromal and cancer cells |
| Intracellular Inhibitors | Extracellular Inhibitors | ||
|---|---|---|---|
| Inhibitors of Protein Kinase(s) | Ab Directed against RTK(s) | ||
| Name | Target(s) | Name | Target(s) |
| Afatinib | EGFR | Cetuximab | Ab anti-EGFR |
| Axitinib | VEGFR | Panitumumab | Ab anti-EGFR |
| Osimertinib | EGFR | Pertuzumab | Ab anti-HER2 |
| Bosutinib | Bcr-Abl, Src | Ramucirumab | Ab anti-VEGF |
| Cabozantinib | MET, AXL, VEGFR, GAS6, RET, ROS1, FLT3, Tie2 | Trastuzumab | Ab anti-HER2 |
| Ceritinib | ALK | Trastuzumab emtansine | Ab anti-HER2 |
| Cobimetinib | MEK | Ab directed against ligand(s) | |
| Crizotinib | ALK and MET | Aflibercept | Ab anti-VEGF |
| Dabrafenib | RAF | Bevabizumab | Ab anti-VEGF |
| Dasatinib | Bcr-Abl, Src | Denosumab | Ab anti-RANKL |
| Erlotinib | EGFR | ||
| Everolimus | mTOR | ||
| Gefitinib | EGFR | ||
| Ibrutinib | BTK | ||
| Idelalisib | p110δ (PI3K) | ||
| Imatinib | Bcr-Abl, c-Kit, DDR1/2, CSF-1R, PDGFR | ||
| Lapatinib | EGFR, ErbB2 | ||
| Lenvatinib | VEGFR, FGFR, PDGFR | ||
| Nilotinib | Bcr-Abl | ||
| Nintedanib | PDGFR, FGFR, VEGFR, FLT3, Lck, Lyn, Src | ||
| Olaparib | PARP | ||
| Osimertinib | EGFR | ||
| Palbociclib | CDK4/6 | ||
| Pazopanib | VEGFR, c-Kit, PDGFR | ||
| Ponatinib | Bcr-Abl | ||
| Regorafenib | VEGFR, c-Kit, PDGFR | ||
| Ribociclib | Cyclin D1/CDK4, CDK6 | ||
| Ruxolitinib | JAK1/2 | ||
| Sonidegib | SMO | ||
| Sorafenib | RAF, VEGFR, FGFR, c-Kit, PDGFR | ||
| Sunitinib | VEGFR, c-Kit, c-Kit, CSF-1R, RET, PDGFR | ||
| Temsirolimus | mTOR | ||
| Tivozanib | VEGF | ||
| Trametinib | MEK1/2 | ||
| Vandetanib | VEGFR, EGFR, RET | ||
| Venetoclax | Bcl2 | ||
| Vemurafenib | ERK, BRAF | ||
| Vismodegib | SMO | ||
| Name of the Study | Molecule Tested | Type of Therapy | Type of Drug | Phase | Pathologies |
|---|---|---|---|---|---|
| D081FC00001-POLO | Olaparib vs. placebo | Targeted therapy | Inhibitor of PARP | III | Metastatic PDAC with BRCA mutation |
| SIRINOX | Oxaliplatin + Irinotecan | Chemotherapy | Platinum salts, DNA topoisomerase I inhibitor | I | Digestive adenocarcinoma (pancreas, oesophagus, stomach, small intestine and biliary tract) |
| PRODIGE 29 | FOLFIRINOX vs. Gemcitabine | Chemotherapy | Anti-metabolite, DNA topoisomerase I inhibitor, Platinum salts | III | Locally advanced PDAC |
| PAMELA-70 | FOLFIRINOX | Chemotherapy | Anti-metabolite, DNA topoisomerase I inhibitor, Platinum salts | II | Metastatic PDAC |
| RC48 | Adoptive transfer of allogeneic lymphocyte cells with natural cytotoxic activity + Cetuximab | Cellular therapy | Antibody anti-EGFR | I/II | Hepatic metastasis of PDAC, colorectal or small intestine cancer |
| PRODIGE 24 − ACCORD 24 | Gemcitabine vs. FOLFIRINOX | Chemotherapy | Anti-metabolite, DNA topoisomerase I inhibitor, Platinum salts | III | PDAC |
| FIRGEMAX | Nab-paclitaxel + Gemcitabine vs. Nab-paclitaxel + Gemcitabine plus FOLFIRI3 | Chemotherapy | Anti-metabolite, DNA topoisomerase I inhibitor, Platinum salts | II | Metastatic PDAC |
| PANOPTIMOX | FOLFIRINOX +/− LV5FU2 vs FOLFIRINOX +/− FIRGEM | Chemotherapy | Anti-metabolite, DNA topoisomerase I inhibitor, Platinum salts | II | Metastatic PDAC |
| MOAnab1 | Gemcitabine + Nab-paclitaxel | Chemotherapy | Anti-metabolite | I | Metastatic PDAC |
| GABRINOX | Gemcitabine + Nab-paclitaxel followed by FOLFIRINOX | Chemotherapy | Anti-metabolite | I | Metastatic PDAC |
| JANUS-2 | Ruxolitinib + Capecitabin | Targeted therapy + Chemotherapy | Inhibitor of Janus kinase (JAK) + Anti-metabolite | III | Locally advanced or metastatic PDAC |
| CMEK162X2111 | MEK162 + Ganitumab | Targeted therapies | MEK inhibitor + antibody anti-IGF1R | I/II | Metastatic PDAC, colorectal adenocarcinoma and melanoma |
| AFUGEM | ABI-007 + Gemcitabine vs. ABI-007 + LV5FU2 | Chemotherapy | Anti-metabolite | II | Metastatic PDAC |
| H9H-MC-JBAJ | Gemcitabine + LY2157299 | Targeted therapy + Chemotherapy | TGFβR inhibitor + Anti-metabolite | I/II | Locally advanced or metastatic PDAC |
| 2009-011992-61 | Gemcitabine + AS703026 | Targeted therapy + Chemotherapy | MEK inhibitor + Anti-metabolite | II | Metastatic PDAC |
| NEOPAC/IPC 2011-002 | Neoadjuvant Gemcitabine + Oxaliplatin and adjuvant Gemcitabine vs. adjuvant Gemcitabine | Chemotherapy | Anti-metabolite +/− Platinum salts | III | PDAC (head of the pancreas) |
| CAOU6 | Gemcitabine +/− ABI-007 | Chemotherapy | Anti-metabolite | III | Metastatic PDAC |
| TherGAP | Anti-tumoural complex CYL-02 | Gene therapy | Enzymatic, Metabolic | I | PDAC |
| ESPAC-4 | Gemcitabine +/− Capecitabin | Chemotherapy | Anti-metabolite | III | Resectable PDAC |
| CO-101-001 | Gemcitabine + CO-1.01 | Chemotherapy | Anti-metabolite | II | Metastatic PDAC |
| GATE 1 | Gemcitabine + Trastuzumab + Erlotinib | Targeted therapies + Chemotherapy | HER2 inhibitor, mTOR inhibitor, Anti-metabolite | II | Metastatic PDAC |
| ASTELLAS 200800 | Gemcitabine + AGS-1C4D4 | Targeted therapy + Chemotherapy | Antibody anti-PSCA + Anti-metabolite | II | Metastatic PDAC |
| AB SCIENCE AB07012 | Gemcitabine +/− Masitinib | Targeted therapy + Chemotherapy | Tyrosine kinase inhibitor + Anti-metabolite | III | Locally advanced or metastatic PDAC |
| THERAPY | Cetuximab + Trastuzumab | Targeted therapies + Chemotherapy | HER2 and EGFR inhibitors | I/II | Metastatic PDAC |
| PANTER | Efavirenz | Targeted therapy | Inhibitor of non-nucleoside reverse transcriptase (NNRTI) | II | PDAC |
| GERCOR LAP 07 D07-1 | Gemcitabine +/− Erlotinib followed by Gemcitabine or chemoradiotherapy with Capecitabin | Targeted therapy + Chemotherapy +/− Chemoradiotherapy | EGFR inhibitor + Anti-metabolite | III | Locally advanced PDAC |
| SciClone SCI-RP-Pan-P2-001 | Gemcitabine +/−RP101 | Targeted therapy + Chemotherapy | Hsp27 inhibitor + Anti-metabolite | II | Unresectable, locally advanced or metastatic PDAC |
| Hoffmann-La Roche BO21129 | Erlotinib | Targeted therapy | EGFR inhibitor | II | Locally advanced PDAC |
| Pharmexa PRIMOVAX | Gemcitabine + GV001 vs. Gemcitabine | Targeted therapy + Chemotherapy | Stimulator of LT CD8 + Anti-metabolite | III | PDAC |
| Hoffmann-La Roche BO21128 | Gemcitabine + Erlotinib | Targeted therapy + Chemotherapy | EGFR inhibitor + Anti-metabolite | II | Metastatic PDAC |
| Sanofi-Aventis EFC10203 | tegafur (a prodrug of 5FU) + gimeracil (5-chloro-2,4 dihydropyridine, CDHP + oteracil (potassium oxonate, Oxo) vs. 5-FU | Chemotherapy | Anti-metabolite | III | Metastatic PDAC |
| Pfizer A4061028 | Gemcitabine +/−AG-013736 (Axitinib) | Targeted therapy + Chemotherapy | VEGFR inhibitor + Anti-metabolite | III | Locally advanced or metastatic unresectable PDAC |
| Sanofi-Aventis EFC10547 | Gemcitabine + Aflibercept | Targeted therapy + Chemotherapy | Antibody anti-VEGF1/2 + Gemcitabine | III | Metastatic PDAC |
| CAPERGEM | Gemcitabine + Capecitabin + Erlotinib | Targeted therapy + Chemotherapy | EGFR inhibitor + Anti-metabolites | I | Advanced PDAC |
| ACCORD 11 PRODIGE 4 | Gemcitabine vs. FOLFIRINOX | Chemotherapy | Anti-metabolite, DNA topoisomerase I inhibitor, Platinum salts | III | Metastatic PDAC |
| ACCORD 09 | Radiotherapy (RT) + Docetaxel + 5-FU or RT + Docetaxel and Cisplatin | Chemoradiotherapy | Radiotherapy, Alkylating agent, Anti-metabolite, Platinum salts | II | PDAC |
| Phase 1-2 (RECF0016) | Radiotherapy + Irinotecan | Chemoradiotherapy | DNA topoisomerase I inhibitor | I/II | Locally advanced PDAC |
| BAYPAN | Gemcitabine +/− Sorafenib | Targeted therapy + Chemotherapy | C-Raf and B-Raf inhibitor + Anti-metabolite | III | Locally advanced or metastatic PDAC |
| Target | Therapy | Number of Patients | Mean Survival (Months) (Treatment Versus Chemo Only) |
|---|---|---|---|
| Telomerase | Gemcitabine + GV1001 | 1062 | 8.4 vs. 6.9 |
| VEGF | Gemcitabine + Bevacizumab | 602 | 5.7 vs. 6.0 |
| Kras | Gemcitabine + Tipifarnib | 688 | 6.3 vs. 6.0 |
| EGFR | Gemcitabine + Cetuximab | 766 | 6.5 vs. 6.0 |
| Gemcitabine + Erlotinib | 569 | 6.24 vs. 5.91 | |
| ErbB2 | Trastuzumab | 44 | 4.6 vs. 5.4 |
| Gastrin | Gastrazol + 5-FU | 98 | 3.6 vs. 4.2 |
| mTOR | Gemcitabine + Everolimus | 29 | 4.5 vs. 6.5 |
| PI3K/PLK | Gemcitabine + Rigosertib | 106 | 6.1 vs. 6.4 |
| Sonic Hedgehog | Gemcitabine + Vismodegib | 106 | 6.9 vs. 6.1 |
| Notch3 | Gemcitabine + IPI-929 | 122 | Not tolerated |
| IGF1-R | Gemcitabine + Ganitumab | 800 | 7.0 vs. 7.2 |
| MMP | Gemcitabine + Matrimastat | 239 | 5.4 vs. 5.4 |
| JAK/STAT | Ruxolitinib + Capecitabin | 127 | 4.5 vs. 4.2 |
| α-secretase | RO4929097 (no Gemcitabine arm) | 18 | 4.1 |
| MEK1/ERK1/2 | Selumetinib versus Capecitabin | 38 | 5.3 vs. 4.9 |
| Name of the Study | Molecule Tested | Type of Therapy | Type of Drug | Type of Patients | Phase |
|---|---|---|---|---|---|
| NCT03065062 | Palbociclib + Gedatolisib | Targeted therapy | CDK4/6 inhibitor + PI3K/mTOR inhibitor | Solid tumors | I |
| NCT02646748 | Pembrolizumab + Itacitinib (INCB039110) and/or Pembrolizumab + INCB050465 | Targeted therapy | PD-1 antibody + JAK1 inhibitor and/or PD-1 antibody + PI3Kδ inhibitor | Solid tumors | I |
| MATCH Screening trial | Multiple (including GSK2636771) | Targeted therapy | Multiple (including PI3Kβ inhibitor) | Advanced refractory solid cancers | II |
| NCT02077933 | Alpelisib + Everolimus or Alpelisib + Everolimus + Exemestane | Targeted therapy | PI3Kα inhibitor + mTOR inhibitor or PI3Kα inhibitor + mTOR inhibitor + aromatase inhibitor | Advanced breast, renal and pancreatic cancer | I |
| NCT02155088 | BYL719 + Gemcitabine + Nab-Paclitaxel | Targeted therapy + chemotherapy | PI3Kα + Anti-metabolite + Microtubule poison | Locally advanced and metastatic pancreatic cancer | I |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cintas, C.; Douché, T.; Therville, N.; Arcucci, S.; Ramos-Delgado, F.; Basset, C.; Thibault, B.; Guillermet-Guibert, J. Signal-Targeted Therapies and Resistance Mechanisms in Pancreatic Cancer: Future Developments Reside in Proteomics. Cancers 2018, 10, 174. https://doi.org/10.3390/cancers10060174
Cintas C, Douché T, Therville N, Arcucci S, Ramos-Delgado F, Basset C, Thibault B, Guillermet-Guibert J. Signal-Targeted Therapies and Resistance Mechanisms in Pancreatic Cancer: Future Developments Reside in Proteomics. Cancers. 2018; 10(6):174. https://doi.org/10.3390/cancers10060174
Chicago/Turabian StyleCintas, Célia, Thibaut Douché, Nicole Therville, Silvia Arcucci, Fernanda Ramos-Delgado, Céline Basset, Benoît Thibault, and Julie Guillermet-Guibert. 2018. "Signal-Targeted Therapies and Resistance Mechanisms in Pancreatic Cancer: Future Developments Reside in Proteomics" Cancers 10, no. 6: 174. https://doi.org/10.3390/cancers10060174






