Hepatic Transcriptome Responses of Domesticated and Wild Turkey Embryos to Aflatoxin B1
Abstract
:1. Introduction
2. Results
2.1. Phenotypic Effects of AFB1
| Type | Exposure | Treatment 1 | Number of Embryos | Egg Weight (g) | Embryo Weight (g) | Liver Weight (g) | Relative Liver Weight (g) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||||
| DT | 1 Day | CNTL | 7 | 79.56 | ±5.62 | 16.58 | ±0.97 | 0.26 | ±0.03 | 1.56 | ±0.12 |
| AFB | 7 | 81.13 | ±8.49 | 17.67 | ±0.49 | 0.28 | ±0.03 | 1.56 | ±0.17 | ||
| 5 Days | CNTL | 7 | 76.6 | ±5.71 | 30.7 | ±1.93 | 0.65 d | ±0.13 | 2.12 f | ±0.37 | |
| AFB | 7 | 82.16 a | ±5.56 | 32.60 b | ±2.86 | 0.53 d | ±0.22 | 1.64 f | ±0.22 | ||
| WT | 1 Day | CNTL | 4 | 66.77 | ±8.75 | 18.71 | ±1.09 | 0.34 | ±0.04 | 1.81 | ±0.20 |
| AFB | 4 | 74.7 | ±4.51 | 19.08 | ±1.17 | 0.33 | ±0.04 | 1.74 | ±0.26 | ||
| 5 Days | CNTL | 3 | 66.3 | ±10.52 | 29.59 c | ±4.46 | 0.64 e | ±0.07 | 2.21 | ±0.34 | |
| AFB | 4 | 60.18 a | ±3.68 | 24.83 b,c | ±0.46 | 0.46 e | ±0.06 | 1.87 | ±0.25 | ||
2.2. RNA-seq Datasets
| Type | Number of Libraries | Read Status | Mean Read Length (bp) | Mean Quality Score | Mean GC Content (%) | Read Pairs | Single Reads | Total Sequence (Gb) |
|---|---|---|---|---|---|---|---|---|
| DT | 24 | Raw | 101.0 | 35.8 | 47.2 | 250,429,631 | NA | 50.6 |
| Corrected | 95.7 | 36.6 | 46.9 | 245,205,648 | 4,829,880 | 47.4 | ||
| WT | 15 | Raw | 101.0 | 34.7 | 46.9 | 190,989,257 | NA | 38.6 |
| Corrected | 95.1 | 35.9 | 46.6 | 184,931,064 | 5,721,404 | 35.7 |
2.3. Mapping to MAKER Gene Set
| Type | Mapped Reads (% of Corrected) 1 | Unmapped Reads (% of Corrected) 1 | Expressed Genes (% of Total Genes) | Mean Read Depth per Gene |
|---|---|---|---|---|
| DT | 380.6 M (76.9%) | 114.6 M (23.1%) | 17,293 (94.7%) | 868 |
| WT | 289.4 M (77.0%) | 86.1 M (22.9%) | 17,211 (94.2%) | 1056 |
| Total | 670.1 M (76.9%) | 200.8 M (23.1%) | 17,440 (95.5%) | 941 |
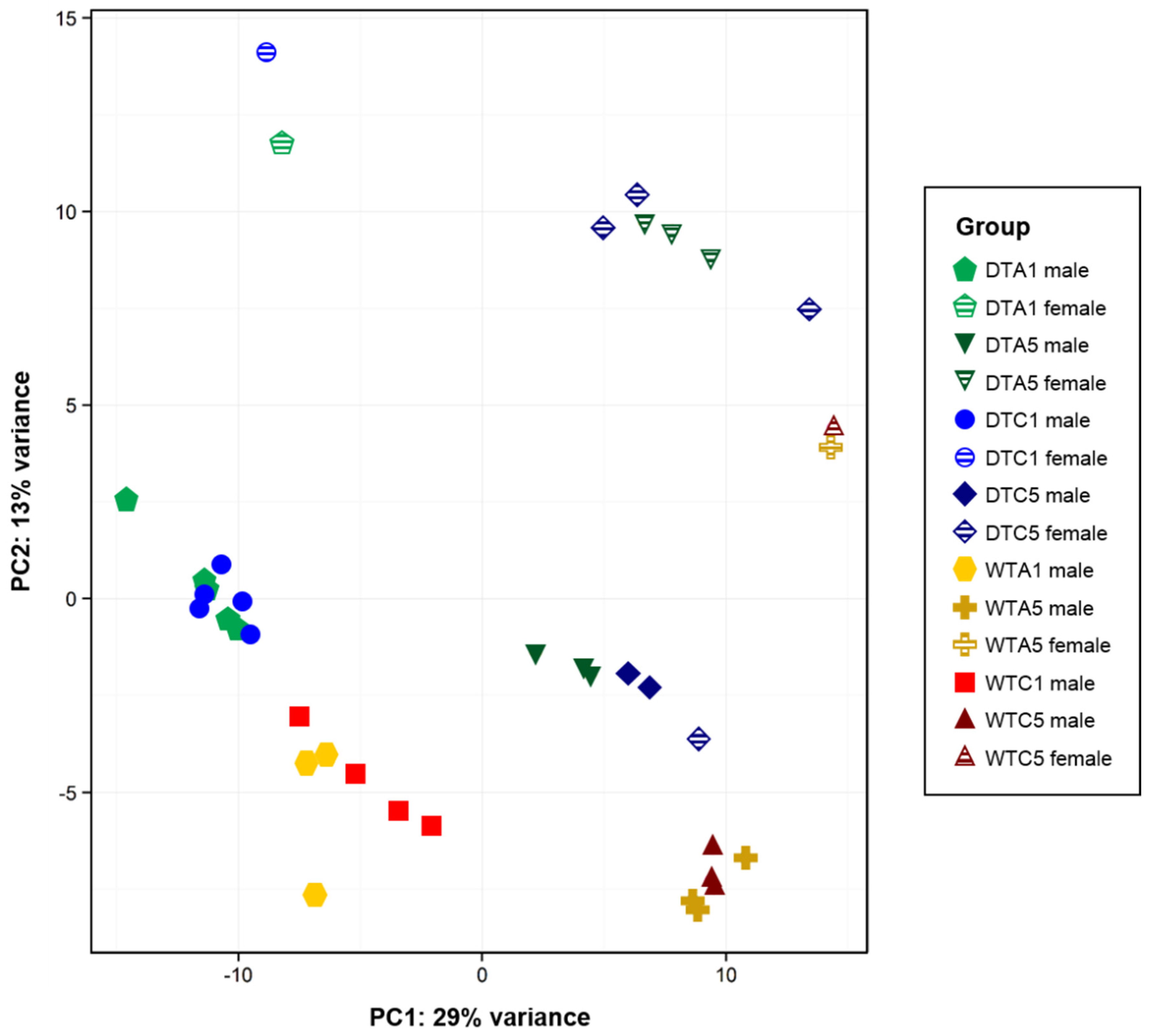
2.4. Sample Variation
2.5. Differential Expression Analysis
2.6. Developmental Effects
2.7. Effects of 1 Day of Exposure to AFB1
| Gene ID | Gene | DTA1/DTC1 | DTA5/DTC5 | WTA1/WTC1 | WTA5/WTC5 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Log2FC | q-value | Log2FC | q-value | Log2FC | q-value | Log2FC | q-value | ||
| T_ALT_S_008999 | AKR1A1 | 0.34 | 3.18 × 10−2 | 0.07 | 7.76 × 10−1 | 0.25 | 8.07 × 10−1 | 0.69 | 5.58 × 10−5 |
| T_ALT_S_014672 | AKR7A2 | 0.47 | 1.15 × 10−2 | 0.14 | 4.91 × 10−1 | 0.34 | 5.55 × 10−1 | 0.18 | 3.37 × 10−1 |
| T_ALT_S_008024 | AOX1 | 0.28 | 3.65 × 10−2 | 0.00 | 9.96 × 10−1 | −0.02 | 9.95 × 10−1 | −0.04 | 8.86 × 10−1 |
| T_ALT_S_003995 | CAT | −0.18 | 3.15 × 10−1 | −0.44 | 3.49 × 10−4 | −0.18 | 8.54 × 10−1 | −0.30 | 1.58 × 10−1 |
| T_ALT_S_010665 | DNAJA3 | 0.40 | 1.15 × 10−3 | −0.03 | 8.79 × 10−1 | 0.07 | 9.78 × 10−1 | 0.21 | 1.66 × 10−1 |
| T_ALT_S_009842 | DNAJB11 | −0.32 | 7.44 × 10−3 | −0.08 | NA | −0.39 | 2.36 × 10−1 | 0.31 | 1.15 × 10−1 |
| T_ALT_S_008078 | DNAJC10 | −0.39 | 1.01 × 10−3 | −0.02 | 9.65 × 10−1 | −0.05 | 9.91 × 10−1 | 0.18 | 4.09 × 10−1 |
| T_ALT_S_014426 | DNAJC11 | 0.26 | 4.81 × 10−2 | −0.06 | 7.63 × 10−1 | −0.20 | 8.47 × 10−1 | 0.20 | 3.88 × 10−1 |
| T_ALT_S_007708 | DNAJC13 | 0.03 | 8.56 × 10−1 | −0.40 | 6.56 × 10−3 | 0.07 | 9.80 × 10−1 | 0.08 | 7.48 × 10−1 |
| T_ALT_S_014061 | DNAJC18 | −0.02 | 8.95 × 10−1 | −0.02 | 9.44 × 10−1 | −0.24 | 5.93 × 10−1 | −0.42 | 1.81 × 10−2 |
| T_ALT_S_002734 | EPHX1 | 0.63 | 5.74 × 10−4 | −0.06 | 8.95 × 10−1 | 0.03 | 9.93 × 10−1 | 0.57 | 2.36 × 10−3 |
| T_ALT_S_003948 | FTH1 | 0.10 | 6.77 × 10−1 | 0.38 | 4.52 × 10−2 | −0.10 | 9.41 × 10−1 | 0.12 | 7.29 × 10−1 |
| T_ALT_S_003347 | GSTA3 1 | 1.60 | 1.47 × 10−10 | 0.74 | 1.79 × 10−2 | 0.48 | 3.63 × 10−1 | 0.52 | 2.34 × 10−1 |
| T_ALT_S_003348 | GSTA3 1 | 1.52 | 2.87 × 10−9 | 0.85 | 4.31 × 10−3 | 0.39 | 5.52 × 10−1 | 0.23 | 6.61 × 10−1 |
| T_ALT_S_003346 | GSTA3 1 | 1.11 | 9.74 × 10−5 | 0.40 | 2.66 × 10−1 | 0.36 | 5.83 × 10−1 | 0.80 | 4.76 × 10−2 |
| T_ALT_S_003345 | GSTA4 | 0.82 | 8.65 × 10−7 | 0.47 | 2.82 × 10−2 | 0.35 | 6.56 × 10−1 | 0.42 | 4.09 × 10−2 |
| T_ALT_S_001023 | GSTK1 | 0.07 | 6.99 × 10−1 | −0.31 | 2.39 × 10−2 | −0.26 | 5.83 × 10−1 | −0.11 | 6.34 × 10−1 |
| T_ALT_S_009475 | GSTO1 | 0.13 | 5.87 × 10−1 | 0.06 | 8.65 × 10−1 | −0.28 | 3.27 × 10−1 | 1.07 | 4.63 × 10−3 |
| T_ALT_S_011992 | HERPUD1 | −0.25 | 3.76 × 10−1 | 0.69 | 3.75 × 10−4 | −0.20 | 9.21 × 10−1 | −0.26 | 3.11 × 10−1 |
| T_ALT_S_000662 | HMOX1 | 0.29 | 1.91 × 10−1 | −0.14 | 7.31 × 10−1 | 0.32 | 3.84 × 10−1 | 0.85 | 1.48 × 10−9 |
| T_ALT_S_005601 | MGST2 | 0.84 | 6.76 × 10−7 | 0.14 | 6.23 × 10−1 | 0.41 | 2.18 × 10−1 | 0.37 | 8.19 × 10−2 |
| T_ALT_S_008700 | MGST3 | 0.56 | 9.79 × 10−5 | 0.10 | 7.83 × 10−1 | 0.33 | 2.55 × 10−1 | 0.73 | 4.64 × 10−4 |
| T_ALT_S_012166 | NQO1 | 0.63 | 2.64 × 10−6 | 0.03 | 8.87 × 10−1 | 0.57 | 1.81 × 10−3 | 0.02 | 9.39 × 10−1 |
| T_ALT_S_004785 | NQO2 | 0.24 | 2.42 × 10−1 | 0.63 | 1.94 × 10−4 | 0.00 | 1.00 | 0.73 | 6.78 × 10−6 |
| T_ALT_S_009000 | PRDX1 | 0.49 | 2.13 × 10−3 | 0.10 | 6.39 × 10−1 | 0.10 | 9.69 × 10−1 | 0.66 | 1.03 × 10−4 |
| T_ALT_S_016734 | STIP1 | −0.03 | 8.93 × 10−1 | −0.19 | 2.64 × 10−1 | −0.14 | 8.98 × 10−1 | 0.36 | 9.48 × 10−3 |
| T_ALT_S_007072 | TXN | 0.09 | 5.73 × 10−1 | −0.62 | 2.26 × 10−3 | −0.05 | 9.80 × 10−1 | −0.07 | 6.80 × 10−1 |
| T_ALT_S_000674 | TXNRD1 1 | 0.61 | 1.32 × 10−2 | 0.15 | 7.63 × 10−1 | 0.54 | 2.55 × 10−1 | 0.45 | 2.82 × 10−1 |
| T_ALT_S_011677 | TXNRD1 1 | −0.30 | 2.07 × 10−3 | 0.00 | 9.99 × 10−1 | 0.00 | 9.99 × 10−1 | −0.16 | 3.06 × 10−1 |
| T_ALT_S_007907 | UGT1A1 | 0.62 | 2.35 × 10−5 | 0.06 | 8.19 × 10−1 | 0.15 | 9.05 × 10−1 | 0.34 | 8.16 × 10−3 |
| T_ALT_S_009396 | MRP2 | 0.16 | 3.07 × 10−1 | −0.32 | 3.68 × 10−2 | 0.24 | 4.82 × 10−1 | 0.24 | 2.59 × 10−1 |
| T_ALT_S_006174 | SOD3 | 0.33 | 3.12 × 10−1 | 0.17 | 7.08 × 10−1 | 0.07 | 9.92 × 10−1 | 0.58 | 2.25 × 10−2 |
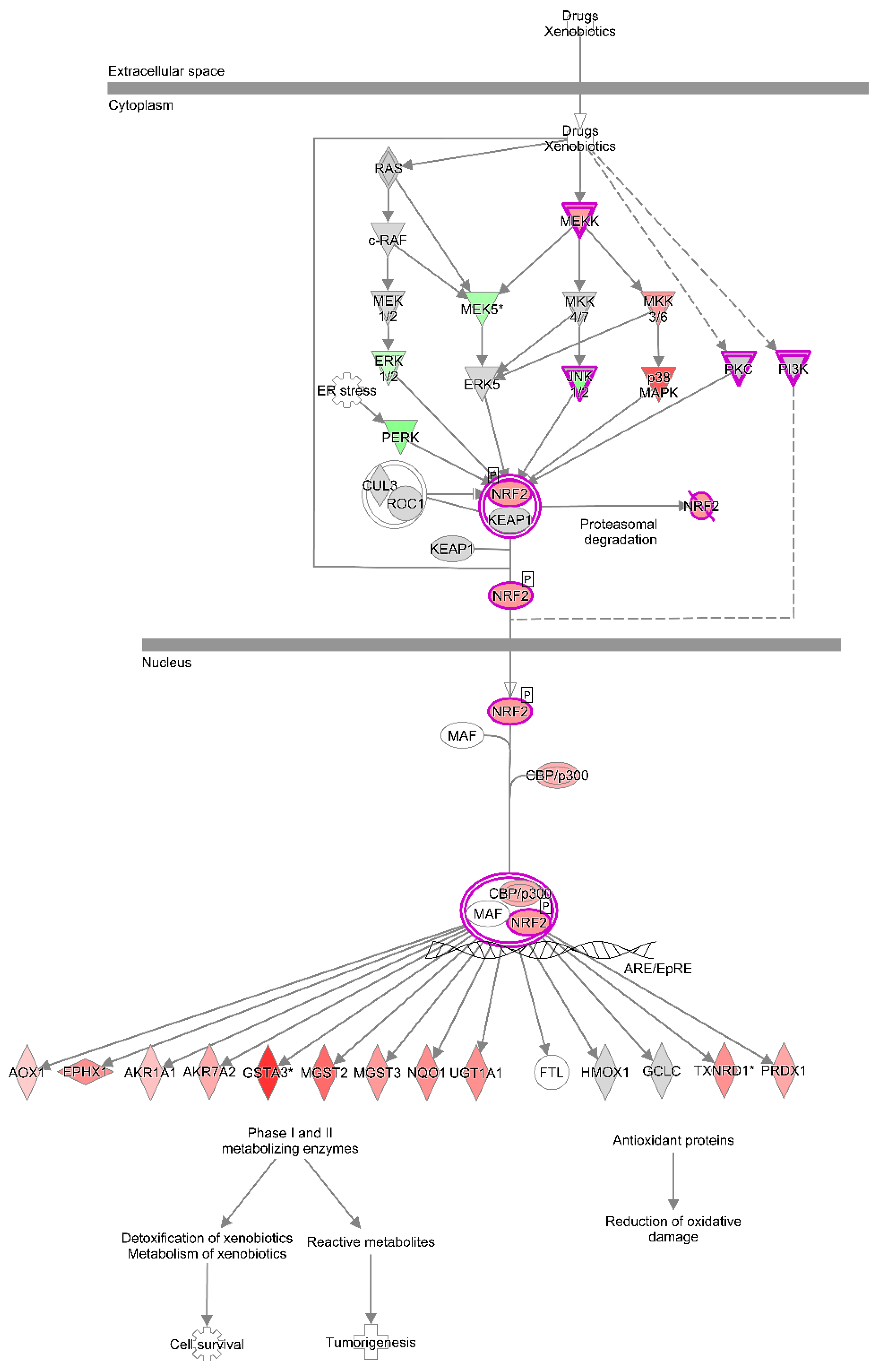
2.8. Effect of 5 Days of Exposure to AFB1
2.9. Gender Effects
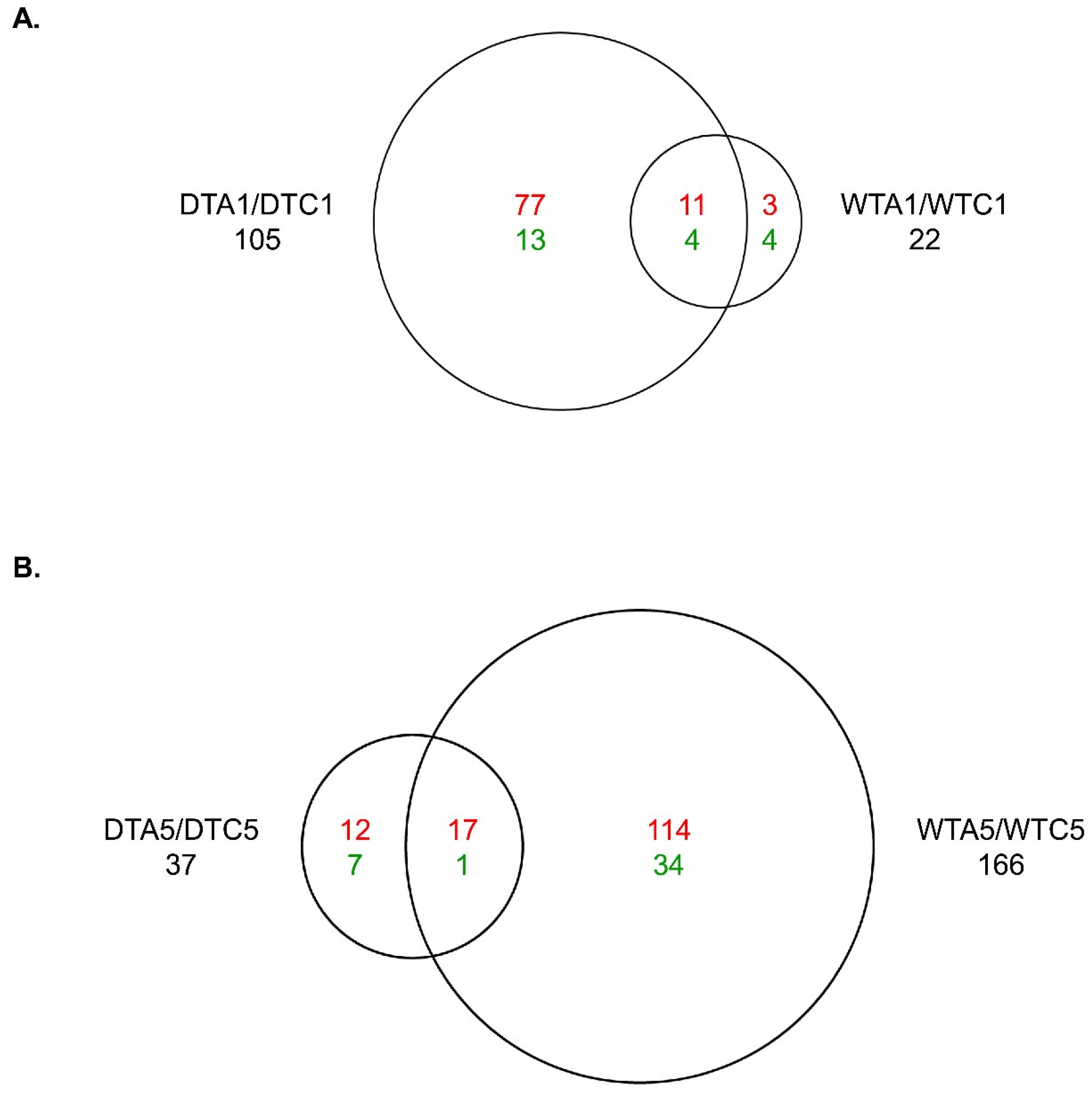
2.10. Comparison of AFB1 Effects

3. Discussion
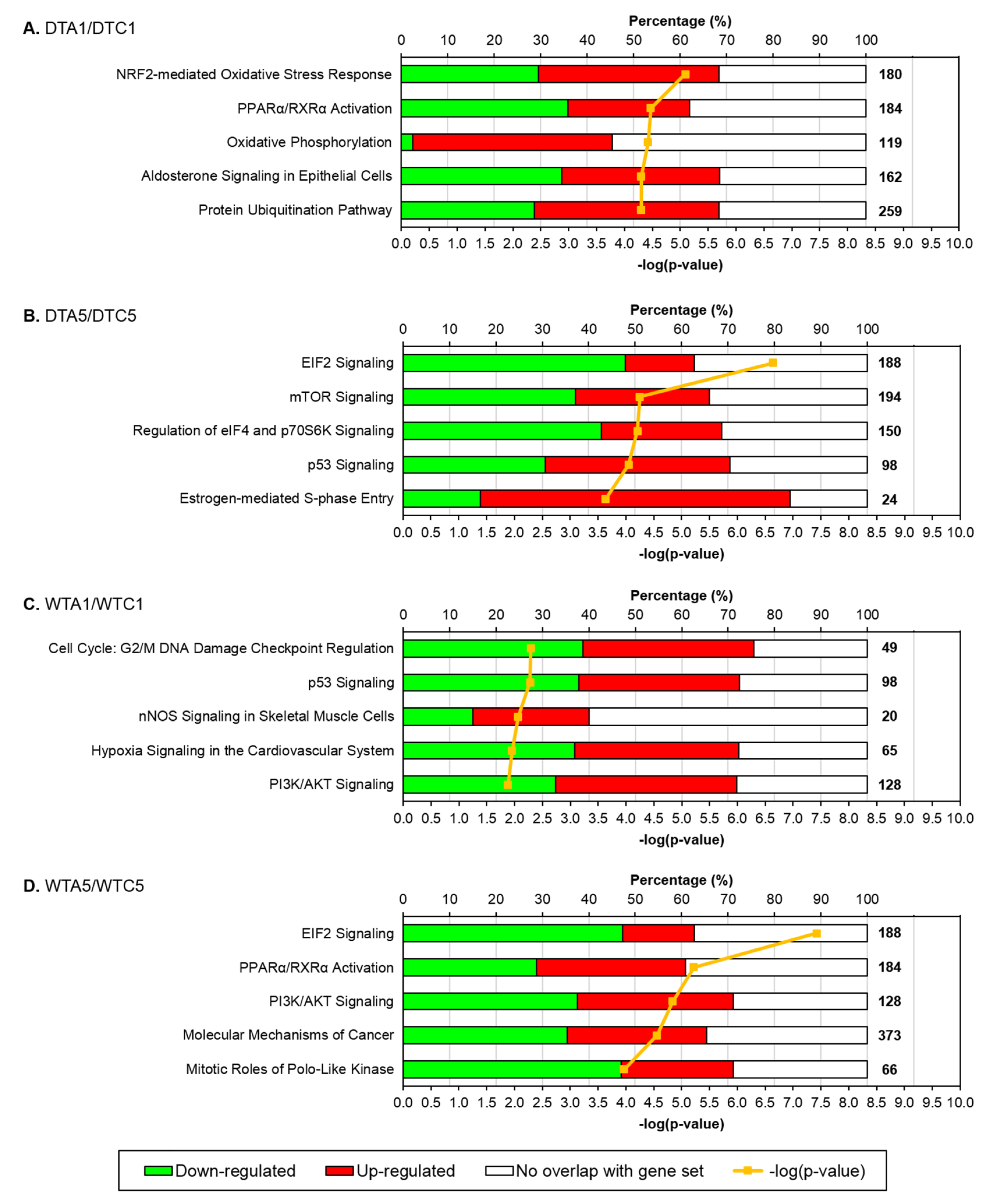
3.1. Responses to AFB1
3.2. Time of Response in WT and DT
3.3. In ovo Exposure as Model of Aflatoxicosis
4. Experimental Section
4.1. Embryos and Toxin Preparation
4.2. In ovo AFB1 Exposure
4.3. RNA Isolation and Sequencing
4.4. Read Filtering, Trimming, and Dataset QC Analysis
4.5. Read Mapping to MAKER Gene Set
| Statistic | Number of Genes per Chromosome 1 | % Annotated Genes per Chromosome 1 | Number of Exons per Gene | Genomic Region (Including Introns) | Exons Only |
|---|---|---|---|---|---|
| Min | 2 | 87.40% | 1 | 6 | 6 |
| Mean | 553 | 93.90% | 10 | 24,805 | 2298 |
| Max | 2228 | 100.00% | 141 | 1,524,779 | 55,896 |
| N50 | N/A | N/A | N/A | 57,428 | 3654 |
| Total | 18,265 | 93.70% | 180,541 | 453,055,619 | 41,981,535 |
4.6. Differential Expression and Functional Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Giambrone, J.J.; Diener, U.L.; Davis, N.D.; Panangala, V.S.; Hoerr, F.J. Effects of aflatoxin on young turkeys and broiler chickens. Poult. Sci. 1985, 64, 1678–1684. [Google Scholar] [CrossRef] [PubMed]
- Pandey, I.; Chauhan, S.S. Studies on production performance and toxin residues in tissues and eggs of layer chickens fed on diets with various concentrations of aflatoxin AFB1. Br. Poult. Sci. 2007, 48, 713–723. [Google Scholar] [CrossRef]
- Rauber, R.H.; Dilkin, P.; Giacomini, L.Z.; Araújo de Almeida, C.A.; Mallmann, C.A. Performance of turkey poults fed different doses of aflatoxins in the diet. Poult. Sci. 2007, 86, 1620–1624. [Google Scholar] [CrossRef] [PubMed]
- Rawal, S.; Kim, J.E.; Coulombe, R., Jr. Aflatoxin B1 in poultry: Toxicology, metabolism and prevention. Res. Vet. Sci. 2010, 89, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Bedard, L.L.; Massey, T.E. Aflatoxin B1-induced DNA damage and its repair. Cancer Lett. 2006, 241, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Eaton, D.L.; Gallagher, E.P. Mechanisms of aflatoxin carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 1994, 34, 135–172. [Google Scholar] [CrossRef] [PubMed]
- Corrier, D.E. Mycotoxicosis: Mechanisms of immunosuppression. Vet. Immunol. Immunopathol. 1991, 30, 73–87. [Google Scholar] [CrossRef]
- Kim, J.E.; Bunderson, B.R.; Croasdell, A.; Coulombe, R.A., Jr. Functional characterization of alpha-class glutathione S-transferases from the turkey (Meleagris gallopavo). Toxicol. Sci. 2011, 124, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Klein, P.J.; Buckner, R.; Kelly, J.; Coulombe, R.A., Jr. Biochemical basis for the extreme sensitivity of turkeys to aflatoxin B1. Toxicol. Appl. Pharmacol. 2000, 165, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Quist, C.F.; Bounous, D.I.; Kilburn, J.V.; Nettles, V.F.; Wyatt, R.D. The effect of dietary aflatoxin on wild turkey poults. J. Wildl. Dis. 2000, 36, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Bunderson, B.R.; Croasdell, A.; Reed, K.M.; Coulombe, R.A., Jr. Alpha-class glutathione S-transferases in wild turkeys (Meleagris gallopavo): Characterization and role in resistance to the carcinogenic mycotoxin aflatoxin B1. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Aly, S.A.; Anwer, W. Effect of naturally contaminated feed with aflatoxins on performance of laying hens and the carryover of aflatoxin B1 residues in table egg. Pakistan J. Nutr. 2009, 8, 181–186. [Google Scholar]
- Azzam, A.H.; Gabal, M.A. Aflatoxin and immunity in layer hens. Avian Pathol. 1998, 27, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Khan, W.A.; Khan, M.Z.; Khan, A.; Hassan, Z.U.; Rafique, S.; Saleemi, M.K.; Ahad, A. Dietary vitamin E in White Leghorn layer breeder hens: A strategy to combat aflatoxin B1-induced damage. Avian Pathol. 2014, 43, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.A.; Kobashigawa, E.; Reis, T.A.; Mestieri, L.; Albuquerque, R.; Corrêa, B. Aflatoxin B1 residues in eggs of laying hens fed a diet containing different levels of the mycotoxin. Food Addit. Contam. 2000, 17, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.A.F.; Rosmaninho, J.F.; Castro, A.L.; Butkeraitis, P.; Alves Reis, T.; Corrêa, B. Aflatoxin residues in eggs of laying Japanese quail after long-term administration of rations containing low levels of aflatoxin B1. Food Addit. Contam. 2003, 20, 648–653. [Google Scholar] [CrossRef] [PubMed]
- Wolzak, A.; Pearson, A.M.; Coleman, T.H.; Pestka, J.J.; Gray, J.I. Aflatoxin deposition and clearance in the eggs of laying hens. Food Chem. Toxicol. 1985, 23, 1057–1061. [Google Scholar] [CrossRef]
- Celik, I.; Oguz, H.; Demet, O.; Boydak, M.; Donmez, H.H.; Sur, E.; Nizamlioglu, F. Embryotoxicity assay of aflatoxin produced by Aspergillus parasiticus NRRL 2999. Br. Poult. Sci. 2000, 41, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Dietert, R.R.; Qureshi, M.A.; Nanna, U.C.; Bloom, S.E. Embryonic exposure to aflatoxin-B1: Mutagenicity and influence on development and immunity. Environ. Mutagen. 1985, 7, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Edrington, T.S.; Harvey, R.B.; Kubena, L.F. Toxic effects of aflatoxin B1 and ochratoxin A, alone and in combination, on chicken embryos. Bull. Environ. Contam. Toxicol. 1995, 54, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Oznurlu, Y.; Celik, I.; Sur, E.; Ozaydın, T.; Oğuz, H.; Altunbaş, K. Determination of the effects of aflatoxin B1 given in ovo on the proximal tibial growth plate of broiler chickens: Histological, histometric and immunohistochemical findings. Avian Pathol. 2012, 41, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, M.A.; Brake, J.; Hamilton, P.B.; Hagler, W.M., Jr.; Nesheim, S. Dietary exposure of broiler breeders to aflatoxin results in immune dysfunction in progeny chicks. Poult. Sci. 1998, 77, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Sur, E.; Celik, I. Effects of aflatoxin B1 on the development of the bursa of Fabricius and blood lymphocyte acid phosphatase of the chicken. Br. Poult. Sci. 2003, 44, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.G.; Deschl, U.; William, G.M. DNA damage in fetal liver cells of turkey and chicken eggs dosed with aflatoxin B1. Arch. Toxicol. 2011, 85, 1167–1172. [Google Scholar] [CrossRef] [PubMed]
- Neldon-Ortiz, D.L.; Qureshi, M.A. Effects of AFB1 embryonic exposure on chicken mononuclear phagocytic cell functions. Dev. Comp. Immunol. 1992, 16, 187–196. [Google Scholar] [CrossRef]
- Sur, E.; Celik, I.; Oznurlu, Y.; Aydin, M.F.; Oguz, H.; Kurtoglu, V.; Ozaydin, T. Enzyme histochemical and serological investigations on the immune system from chickens treated in ovo with aflatoxin B1 (AFB1). Rev. Méd. Vét. 2011, 162, 443–448. [Google Scholar]
- Ul-Hassan, Z.; Khan, M.Z.; Khan, A.; Javed, I. Immunological status of the progeny of breeder hens kept on ochratoxin A (OTA)- and aflatoxin B1 (AFB1)-contaminated feeds. J. Immunotoxicol. 2012, 9, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Monson, M.S.; Settlage, R.E.; McMahon, K.W.; Mendoza, K.M.; Rawal, S.; El-Nezami, H.S.; Coulombe, R.A.; Reed, K.M. Response of the hepatic transcriptome to aflatoxin B1 in domestic turkey (Meleagris gallopavo). PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monson, M.S.; Settlage, R.E.; Mendoza, K.M.; Rawal, S.; El-Nezami, H.S.; Coulombe, R.A.; Reed, K.M. Modulation of the spleen transcriptome in domestic turkey (Meleagris gallopavo) in response to aflatoxin B1 and probiotics. Immunogenetics 2015, 67, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Bauer, M.M.; Mendoza, K.M.; Reed, K.M.; Coulombe, R.A., Jr. Comparative genomics identifies new alpha class genes within the avian glutathione S-transferase gene cluster. Gene 2010, 452, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Horn, N.; Applegate, T.J. Efficiency of hydrated sodium calcium aluminosilicate to ameliorate the adverse effects of graded levels of aflatoxin B1 in broiler chicks. Poult. Sci. 2014, 93, 2037–2047. [Google Scholar] [CrossRef] [PubMed]
- Giambrone, J.J.; Diener, U.L.; Davis, N.D.; Panangala, V.S.; Hoerr, F.J. Effects of purified aflatoxin on broiler chickens. Poult. Sci. 1985, 64, 852–858. [Google Scholar] [CrossRef] [PubMed]
- Giambrone, J.J.; Diener, U.L.; Davis, N.D.; Panangala, V.S.; Hoerr, F.J. Effects of purified aflatoxin on turkeys. Poult. Sci. 1985, 64, 859–865. [Google Scholar] [CrossRef] [PubMed]
- Huff, W.E.; Kubena, L.F.; Harvey, R.B.; Corrier, D.E.; Mollenhauer, H.H. Progression of aflatoxicosis in broiler chickens. Poult. Sci. 1986, 65, 1891–1899. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.A.; Rosmaninho, J.F.; Butkeraitis, P.; Corrêa, B.; Reis, T.A.; Guerra, J.L.; Albuquerque, R.; Moro, M.E. Effect of low levels of dietary aflatoxin B1 on laying Japanese quail. Poult. Sci. 2002, 81, 976–980. [Google Scholar] [CrossRef] [PubMed]
- Verma, J.; Johri, T.S.; Swain, B.K. Effect of varying levels of aflatoxin, ochratoxin and their combinations on the performance and egg quality characteristics in laying hens. Asian Aust. J. Anim. Sci. 2003, 16, 1015–1019. [Google Scholar] [CrossRef]
- Yarru, L.P.; Settivari, R.S.; Antoniou, E.; Ledoux, D.R.; Rottinghaus, G.E. Toxicological and gene expression analysis of the impact of aflatoxin B1 on hepatic function of male broiler chicks. Poult. Sci. 2009, 88, 360–371. [Google Scholar] [CrossRef] [PubMed]
- Yarru, L.P.; Settivari, R.S.; Gowda, N.K.; Antoniou, E.; Ledoux, D.R.; Rottinghaus, G.E. Effects of turmeric (Curcuma longa) on the expression of hepatic genes associated with biotransformation, antioxidant, and immune systems in broiler chicks fed aflatoxin. Poult. Sci. 2009, 88, 2620–2627. [Google Scholar] [CrossRef] [PubMed]
- Jennen, D.G.; Magkoufopoulou, C.; Ketelslegers, H.B.; van Herwijnen, M.H.; Kleinjans, J.C.; van Delft, J.H. Comparison of HepG2 and HepaRG by whole-genome gene expression analysis for the purpose of chemical hazard identification. Toxicol. Sci. 2010, 115, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Merrick, B.A.; Phadke, D.P.; Auerbach, S.S.; Mav, D.; Stiegelmeyer, S.M.; Shah, R.R.; Tice, R.R. RNA-Seq profiling reveals novel hepatic gene expression pattern in aflatoxin B1 treated rats. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Rustemeyer, S.M.; Lamberson, W.R.; Ledoux, D.R.; Wells, K.; Austin, K.J.; Cammack, K.M. Effects of dietary aflatoxin on the hepatic expression of apoptosis genes in growing barrows. J. Anim. Sci. 2011, 89, 916–925. [Google Scholar] [CrossRef] [PubMed]
- Josse, R.; Dumont, J.; Fautrel, A.; Robin, M.A.; Guillouzo, A. Identification of early target genes of aflatoxin B1 in human hepatocytes, inter-individual variability and comparison with other genotoxic compounds. Toxicol. Appl. Pharmacol. 2012, 258, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Ricordy, R.; Gensabella, G.; Cacci, E.; Augusti-Tocco, G. Impairment of cell cycle progression by aflatoxin B1 in human cell lines. Mutagenesis 2002, 17, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Ranchal, I.; González, R.; Bello, R.I.; Ferrín, G.; Hidalgo, A.B.; Linares, C.I.; Aguilar-Melero, P.; González-Rubio, S.; Barrera, P.; Marchal, T.; et al. The reduction of cell death and proliferation by p27Kip1 minimizes DNA damage in an experimental model of genotoxicity. Int. J. Cancer 2009, 125, 2270–2280. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Fang, J.; Peng, X.; Cui, H.; Chen, J.; Wang, F.; Chen, Z.; Zuo, Z.; Deng, J.; Lai, W.; Zhou, Y. Effect of selenium supplementation on aflatoxin B1-induced histopathological lesions and apoptosis in bursa of Fabricius in broilers. Food Chem. Toxicol. 2014, 74, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Cova, L.; Wild, C.P.; Mehrotra, R.; Turusov, V.; Shirai, T.; Lambert, V.; Jacquet, C.; Tomatis, L.; Trépo, C.; Montesano, R. Contribution of aflatoxin B1 and hepatitis B virus infection in the induction of liver tumors in ducks. Cancer Res. 1990, 50, 2156–2163. [Google Scholar] [PubMed]
- Cullen, J.M.; Marion, P.L.; Sherman, G.J.; Hong, X.; Newbold, J.E. Hepatic neoplasms in aflatoxin B1-treated, congenital duck hepatitis B virus-infected, and virus-free Pekin ducks. Cancer Res. 1990, 50, 4072–4080. [Google Scholar] [PubMed]
- Klein, P.J.; Van Vleet, T.R.; Hall, J.O.; Coulombe, R.A., Jr. Dietary butylated hydroxytoluene protects against aflatoxicosis in turkeys. Toxicol. Appl. Pharmacol. 2002, 182, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Haupt, Y.; Maya, R.; Kazaz, A.; Oren, M. Mdm2 promotes the rapid degradation of p53. Nature 1997, 387, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.S.; Desterro, J.M.; Lain, S.; Lane, D.P.; Hay, R.T. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol. Cell Biol. 2000, 20, 8458–8467. [Google Scholar] [CrossRef] [PubMed]
- García-Trevijano, E.R.; Latasa, M.U.; Carretero, M.V.; Berasain, C.; Mato, J.M.; Avila, M.A. S-adenosylmethionine regulates MAT1A and MAT2A gene expression in cultured rat hepatocytes: A new role for S-adenosylmethionine in the maintenance of the differentiated status of the liver. FASEB J. 2000, 14, 2511–2518. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.C.; Mato, J.M. S-Adenosylmethionine in cell growth, apoptosis and liver cancer. J. Gastroenterol. Hepatol. 2008, 23, S73–S77. [Google Scholar] [CrossRef] [PubMed]
- Mato, J.M.; Martínez-Chantar, M.L.; Lu, S.C. S-adenosylmethionine metabolism and liver disease. Ann. Hepatol. 2013, 12, 183–189. [Google Scholar] [PubMed]
- Kansanen, E.; Kuosmanen, S.M.; Leinonen, H.; Levonen, A.L. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol. 2013, 1, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Niture, S.K.; Khatri, R.; Jaiswal, A.K. Regulation of Nrf2—An update. Free Radic. Biol. Med. 2014, 66, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Apopa, P.L.; He, X.; Ma, Q. Phosphorylation of Nrf2 in the transcription activation domain by casein kinase 2 (CK2) is critical for the nuclear translocation and transcription activation function of Nrf2 in IMR-32 neuroblastoma cells. J. Biochem. Mol. Toxicol. 2008, 22, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Jowsey, I.R.; Jiang, Q.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Expression of the aflatoxin B1-8,9-epoxide-metabolizing murine glutathione S-transferase A3 subunit is regulated by the Nrf2 transcription factor through an antioxidant response element. Mol. Pharmacol. 2003, 64, 1018–1028. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Sun, Z.; Wang, X.J.; Jiang, T.; Huang, Z.; Fang, D.; Zhang, D.D. Direct interaction between Nrf2 and p21Cip1/WAF1 upregulates the Nrf2-mediated antioxidant response. Mol. Cell. 2009, 34, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Judah, D.J.; Neal, G.E.; Nguyen, T. Molecular cloning and heterologous expression of a cDNA encoding a mouse glutathione S-transferase Yc subunit possessing high catalytic activity for aflatoxin B1-8,9-epoxide. Biochem. J. 1992, 285 Pt 1, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Ilic, Z.; Crawford, D.; Vakharia, D.; Egner, P.A.; Sell, S. Glutathione-S-transferase A3 knockout mice are sensitive to acute cytotoxic and genotoxic effects of aflatoxin B1. Toxicol. Appl. Pharmacol. 2010, 242, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Perrone, C.E.; Ahr, H.J.; Duan, J.D.; Jeffrey, A.M.; Schmidt, U.; Williams, G.M.; Enzmann, H.H. Embryonic turkey liver: Activities of biotransformation enzymes and activation of DNA-reactive carcinogens. Arch. Toxicol. 2004, 78, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, K.; Yuan, S.; Peng, X.; Fang, J.; Wang, F.; Cui, H.; Chen, Z.; Yuan, J.; Geng, Y. Effects of aflatoxin B1 on oxidative stress markers and apoptosis of spleens in broilers. Toxicol. Ind. Health 2013. [Google Scholar] [CrossRef] [PubMed]
- Eraslan, G.; Akdogan, M.; Yarsan, E.; Sahindokuyucu, F.; Essiz, D.; Altintas, L. The effects of aflatoxins on oxidative stress in broiler chickens. Turk J. Vet. Anim. Sci. 2005, 29, 701–707. [Google Scholar]
- Shen, H.M.; Shi, C.Y.; Lee, H.P.; Ong, C.N. Aflatoxin B1-induced lipid peroxidation in rat liver. Toxicol. Appl. Pharmacol. 1994, 127, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P.; Cai, H.; McMahon, M.; Hayes, J.D.; Sutter, T.R.; Groopman, J.D.; Deng, Z.; Harris, T.M. Reduction of aflatoxin B1 dialdehyde by rat and human aldo-keto reductases. Chem. Res. Toxicol. 2001, 14, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Judah, D.J.; Hayes, J.D.; Yang, J.C.; Lian, L.Y.; Roberts, G.C.; Farmer, P.B.; Lamb, J.H.; Neal, G.E. A novel aldehyde reductase with activity towards a metabolite of aflatoxin B1 is expressed in rat liver during carcinogenesis and following the administration of an anti-oxidant. Biochem. J. 1993, 292, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Kelly, E.J.; Erickson, K.E.; Sengstag, C.; Eaton, D.L. Expression of human microsomal epoxide hydrolase in Saccharomyces cerevisiae reveals a functional role in aflatoxin B1 detoxification. Toxicol. Sci. 2002, 65, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Klein, P.J.; Van Vleet, T.R.; Hall, J.O.; Coulombe, R.A., Jr. Biochemical factors underlying the age-related sensitivity of turkeys to aflatoxin B1. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2002, 132, 193–201. [Google Scholar] [CrossRef]
- Klein, P.J.; Van Vleet, T.R.; Hall, J.O.; Coulombe, R.A., Jr. Effects of dietary butylated hydroxytoluene on aflatoxin B1-relevant metabolic enzymes in turkeys. Food Chem. Toxicol. 2003, 41, 671–678. [Google Scholar] [CrossRef]
- Jaiswal, A.K. Regulation of genes encoding NAD(P)H:quinone oxidoreductases. Free Radic. Biol. Med. 2000, 29, 254–262. [Google Scholar] [CrossRef]
- Toyomizu, M.; Kikusato, M.; Kawabata, Y.; Azad, M.A.; Inui, E.; Amo, T. Meat-type chickens have a higher efficiency of mitochondrial oxidative phosphorylation than laying-type chickens. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2011, 159, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Bottje, W.; Pumford, N.R.; Ojano-Dirain, C.; Iqbal, M.; Lassiter, K. Feed efficiency and mitochondrial function. Poult. Sci. 2006, 85, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Ramachandra Pai, M.; Jayanthi Bai, N.; Venkitasubramanian, T.A. Effect of aflatoxins on oxidative phosphorylation by rat liver mitochondria. Chem. Biol. Interact. 1975, 10, 123–131. [Google Scholar] [PubMed]
- Sajan, M.P.; Satav, J.G.; Bhattacharya, R.K. Activity of some respiratory enzymes and cytochrome contents in rat hepatic mitochondria following aflatoxin B1 administration. Toxicol. Lett. 1995, 80, 55–60. [Google Scholar] [CrossRef]
- Rawal, S.; Yip, S.S.; Coulombe, R.A., Jr. Cloning, expression and functional characterization of cytochrome P450 3A37 from turkey liver with high aflatoxin B1 epoxidation activity. Chem. Res. Toxicol. 2010, 23, 1322–1329. [Google Scholar] [CrossRef] [PubMed]
- Rawal, S.; Coulombe, R.A., Jr. Metabolism of aflatoxin B1 in turkey liver microsomes: The relative roles of cytochromes P450 1A5 and 3A37. Toxicol. Appl. Pharmacol. 2011, 254, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Reed, K.M.; Mendoza, K.M.; Coulombe, R.A., Jr. Structure and genetic mapping of the cytochrome P450 gene (CYP1A5) in the turkey (Meleagris gallopavo). Cytogenet. Genome Res. 2007, 116, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.S.; Coulombe, R.A., Jr. Molecular cloning and expression of a novel cytochrome P450 from turkey liver with aflatoxin B1 oxidizing activity. Chem. Res. Toxicol. 2006, 19, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Monson, M.S. Hepatotoxic and Immunomodulatory Transcriptome Responses to Aflatoxin B1 in the Turkey (Meleagris gallopavo). Ph.D. Thesis, University of Minnesota, St. Paul, MN, USA, 2015. [Google Scholar]
- Friedman, A.; Elad, O.; Cohen, I.; Bar Shira, E. The gut associated lymphoid system in the post-hatch chick: Dynamics of maternal IgA. Israel J. Vet. Med. 2012, 67, 75–81. [Google Scholar]
- Hamal, K.R.; Burgess, S.C.; Pevzner, I.Y.; Erf, G.F. Maternal antibody transfer from dams to their egg yolks, egg whites, and chicks in meat lines of chickens. Poult. Sci. 2006, 85, 1364–1372. [Google Scholar] [CrossRef] [PubMed]
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous inference in general parametric models. Biom J. 2008, 50, 346–363. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2014. Available online: http://www.R-project.org/ (accessed on 4 January 2016).
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 6 October 2011).
- Campbell, M.; (University of Utah, Salt Lake City, UT, USA); Yandell, M.; (University of Utah, Salt Lake City, UT, USA). Personal communication, 2015.
- Cantarel, B.L.; Korf, I.; Robb, S.M.; Parra, G.; Ross, E.; Moore, B.; Holt, C.; Sánchez Alvarado, A.; Yandell, M. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008, 18, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Settlage, R.; (Virginia Tech, Blacksburg, VA, USA). Personal communication, 2015.
- Granevitze, Z.; Blum, S.; Cheng, H.; Vignal, A.; Morisson, M.; Ben-Ari, G.; David, L.; Feldman, M.W.; Weigend, S.; Hillel, J. Female-specific DNA sequences in the chicken genome. J. Hered. 2007, 98, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Kalina, J.; Mucksová, J.; Yan, H.; Trefil, P. Rapid sexing of selected galliformes by polymerase chain reaction. Czech J. Anim. Sci. 2012, 57, 187–192. [Google Scholar]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monson, M.S.; Cardona, C.J.; Coulombe, R.A.; Reed, K.M. Hepatic Transcriptome Responses of Domesticated and Wild Turkey Embryos to Aflatoxin B1. Toxins 2016, 8, 16. https://doi.org/10.3390/toxins8010016
Monson MS, Cardona CJ, Coulombe RA, Reed KM. Hepatic Transcriptome Responses of Domesticated and Wild Turkey Embryos to Aflatoxin B1. Toxins. 2016; 8(1):16. https://doi.org/10.3390/toxins8010016
Chicago/Turabian StyleMonson, Melissa S., Carol J. Cardona, Roger A. Coulombe, and Kent M. Reed. 2016. "Hepatic Transcriptome Responses of Domesticated and Wild Turkey Embryos to Aflatoxin B1" Toxins 8, no. 1: 16. https://doi.org/10.3390/toxins8010016