Draft Genome Sequences of Two Bacillus thuringiensis Strains and Characterization of a Putative 41.9-kDa Insecticidal Toxin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Plasmids
2.2. DNA Isolation, Sequencing and Computational Analysis
2.3. Amplification and Cloning of the Full-Length 41.9-kDa-Protein Gene
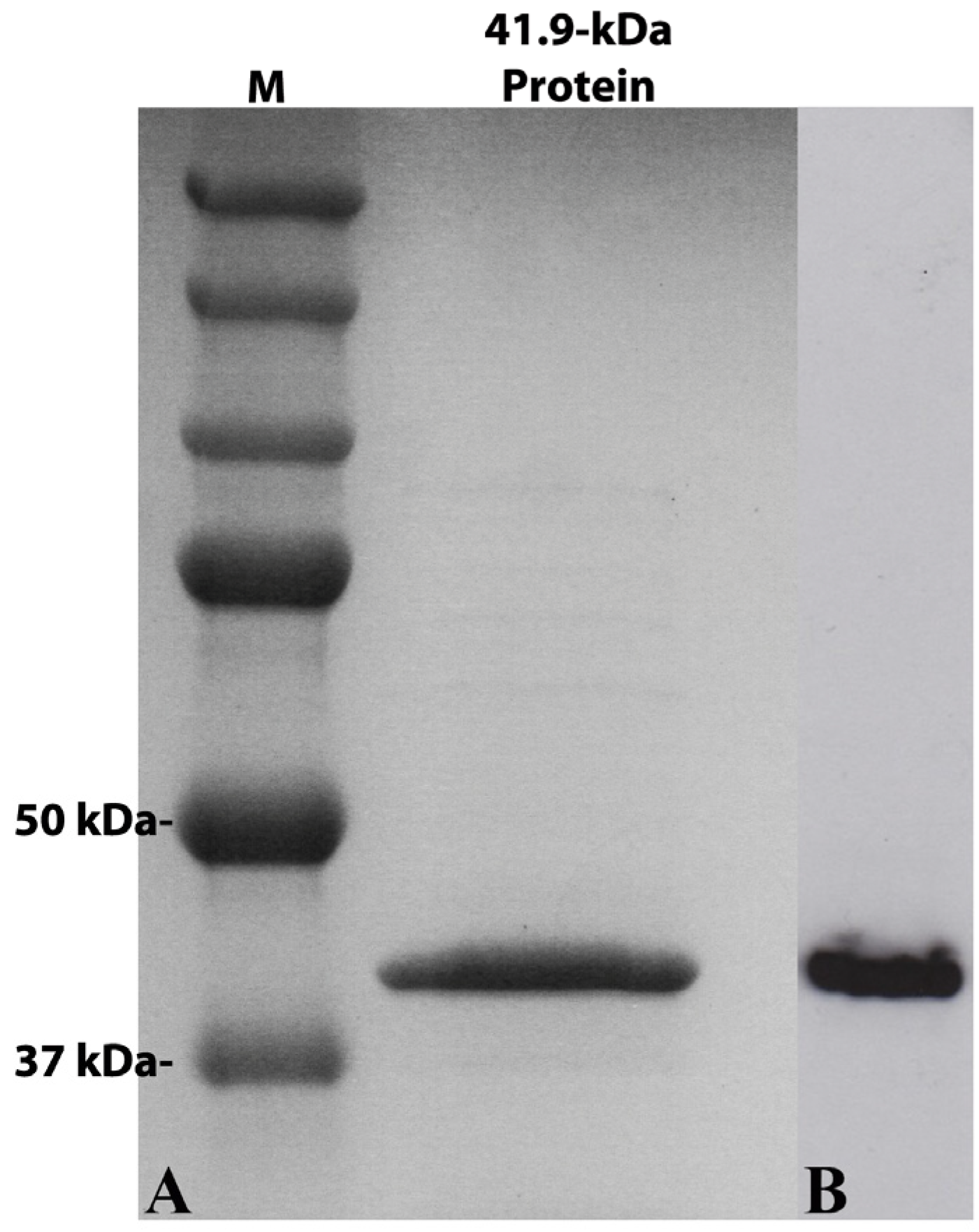
2.4. Expression and Purification of the 41.9-kDa Recombinant Protein
2.5. Insect Rearing and Bioassays
2.6. Nucleotide Sequences Accession Numbers
3. Results and Discussion
3.1. Genome Assembly and Annotation of Bt Strains Hu4-2 and Leapi01
| Strain | Closest homolog | Accesion number | % aa identity | CDS completeness |
|---|---|---|---|---|
| Hu4-2 | Cry1Ad | AAA22340 | 100 | Incomplete |
| Vip3Ba | AAV70653 | 46 | VTGR | |
| Cry9Ea | BAA34908 | 100 | Full-length | |
| Cry1Ab | AAA22330 | 100 | Incomplete | |
| Cry1Ad | AAA22340 | 100 | Incomplete | |
| Cry1Fa | AAA22348 | 100 | Incomplete | |
| Vip3Ca2 | AEE98106 | 100 | Full-length | |
| 41.9 kDa | ZP_04231421 | 62 | Full-length | |
| Cry1Da | CAA38099 | 100 | Incomplete | |
| Cry1Ia7 | AAM73516 | 100 | Full-length | |
| Cry1Ca+P19 | CAA30396 | 100 | Incomplete | |
| Cry1Aa | AAA22353 | 100 | Incomplete | |
| Cry1Ca | CAA30396 | 100 | Incomplete | |
| Cry1Fa | AAA22348 | 100 | Incomplete | |
| Cry1Fa | AAA22348 | 100 | Incomplete | |
| Cry1Ea | CAA37933 | 99 | Incomplete | |
| Cry1Da | CAA38099 | 100 | Incomplete | |
| Cry2Ab | ACC86136 | 98 | VTGR | |
| Cry1Fa+P19 | AAA22348 | 100 | Incomplete | |
| Cry1Ab | AAA22330 | 100 | Incomplete | |
| Cry1Ad | Q03744 | 99 | Incomplete | |
| Cry1Fa | AAA22348 | 100 | Incomplete | |
| Leapi01 | Cry1Aa | AAA22330 | 99 | Incomplete |
| Cry1Ab | AAA22330 | 100 | Incomplete | |
| Cry9Ea | CAA85764 | 100 | Full-length | |
| Cry1Ca | CAA30396 | 100 | Full-length * | |
| Cry1Da | CAA38099 | 100 | Full-length * | |
| Cry1Ia2 | CAA44633 | 100 | Full-length | |
| Cry1Ab | AAA22330 | 100 | VTGR | |
| Cry1Aa | AAA22353 | 100 | Incomplete | |
| Cry1Ab | AAA22330 | 100 | Incomplete | |
| 41.9-kDa | ZP_04231421 | 62 | Full-length | |
| Vip3Aa10 | AAC37036 | 99 | Full-length | |
| Cry2Ab | AAA22342 | 100 | Full-length | |
| Cry1Aa | BAA00257 | 100 | Full-length ** | |
| Cry1Ia14 | ACG63871 | 100 | Full-length ** |
3.2. Molecular Characterization of the Putative 41.9-kDa Insecticidal Protein
3.3. Insecticidal Bioassay of the Putative 41-kDa Insecticidal Protein
4. Conclusions
Acknowledgments
Authors Contribution
Conflicts of Interest
References
- Schnepf, E.; Crickmore, N.; van Rie, J.; Lereclus, D.; Baum, J.; Feitelson, J.; Zeigler, D.R.; Dean, D.H. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 1998, 62, 775–806. [Google Scholar]
- Van Frankenhuyzen, K. Insecticidal activity of Bacillus thuringiensis crystal proteins. J. Invertebr. Pathol. 2009, 101, 1–16. [Google Scholar] [CrossRef]
- Wei, J.Z.; Hale, K.; Carta, L.; Platzer, E.; Wong, C.; Fang, S.C.; Aroian, R.V. Bacillus thuringiensis crystal proteins that target nematodes. Proc. Natl. Acad. Sci. USA 2003, 100, 2760–2765. [Google Scholar]
- Kondo, S.; Mizuki, E.; Akao, T.; Ohba, M. Antitrichomonal strains of Bacillus thuringiensis. Parasitol. Res. 2002, 88, 1090–1092. [Google Scholar] [CrossRef]
- Ohba, M.; Mizuki, E.; Uemori, A. Parasporin, a new anticancer protein group from Bacillus thuringiensis. Anticancer Res. 2009, 29, 427–433. [Google Scholar]
- Okumura, S.; Ohba, M.; Mizuki, E.; Crickmore, N.; Coté, J.-C.; Nagamatsu, Y.; Kitada, S.; Sakai, H.; Harata, K.; Shin, T. Parasporin nomenclature. Available online: http://parasporin.fitc.pref.fukuoka.jp/ (accessed on 03 February 2014).
- Agaisse, H.; Lereclus, D. How does Bacillus thuringiensis produce so much insecticidal crystal protein? J. Bacteriol. 1995, 177, 6027–6032. [Google Scholar]
- Manasherob, R.; Zaritsky, A.; Ben-Dov, E.; Saxena, D.; Barak, Z.; Einav, M. Effect of accessory proteins P19 and P20 on cytolytic activity of Cyt1Aa from Bacillus thuringiensis subsp. israelensis in Escherichia coli. Curr. Microbiol. 2001, 43, 355–364. [Google Scholar] [CrossRef]
- Shao, Z.; Liu, Z.; Yu, Z. Effects of the 20-kilodalton helper protein on Cry1Ac production and spore formation in Bacillus thuringiensis. Appl. Environ. Microbiol. 2001, 67, 5362–5369. [Google Scholar] [CrossRef]
- Xu, Y.; Nagai, M.; Bagdasarian, M.; Smith, T.W.; Walker, E.D. Expression of the p20 gene from Bacillus thuringiensis H-14 increases Cry11A toxin production and enhances mosquito-larvicidal activity in recombinant gram-negative bacteria. Appl. Environ. Microbiol. 2001, 67, 3010–3015. [Google Scholar] [CrossRef]
- Estruch, J.J.; Warren, G.W.; Mullins, M.A.; Nye, G.J.; Craig, J.A.; Koziel, M.G. Vip3A, a novel Bacillus thuringiensis vegetative insecticidal protein with a wide spectrum of activities against lepidopteran insects. Proc. Natl. Acad. Sci. USA 1996, 93, 5389–5394. [Google Scholar] [CrossRef]
- Warren, G.W.; Koziel, M.G.; Mullins, M.A.; Nye, G.J.; Carr, B.; Desai, N.M.; Kostichka, K.; Duck, N.B.; Estruch, J.J. Auxiliary proteins for enhancing the insecticidal activity of pesticidal proteins. US Patent 5770696, 23 June 1998. [Google Scholar]
- Sattar, S.; Maiti, M.K. Molecular characterization of a novel vegetative insecticidal protein from Bacillus thuringiensis effective against sap-sucking insect pest. J. Microbiol. Biotechnol. 2011, 21, 937–946. [Google Scholar] [CrossRef]
- Donovan, W.P.; Engleman, J.T.; Donovan, J.C.; Baum, J.A.; Bunkers, G.J.; Chi, D.J.; Clinton, W.P.; English, L.; Heck, G.R.; Ilagan, O.M.; et al. Discovery and characterization of Sip1A: A novel secreted protein from Bacillus thuringiensis with activity against coleopteran larvae. Appl. Microbiol. Biotechnol. 2006, 72, 713–719. [Google Scholar] [CrossRef]
- Berry, C. The bacterium, Lysinibacillus sphaericus, as an insect pathogen. J. Invertebr. Pathol. 2012, 109, 1–10. [Google Scholar] [CrossRef]
- Peña, G.; Miranda-Rios, J.; de la Riva, G.; Pardo-López, L.; Soberon, M.; Bravo, A. A Bacillus thuringiensis S-layer protein involved in toxicity against Epilachna varivestis (Coleoptera: Coccinellidae). Appl. Environ. Microbiol. 2006, 72, 353–360. [Google Scholar] [CrossRef]
- Raymond, B.; Johnston, P.R.; Nielsen-LeRoux, C.; Lereclus, D.; Crickmore, N. Bacillus thuringiensis: An impotent pathogen? Trends Microbiol. 2010, 18, 189–194. [Google Scholar] [CrossRef]
- Mac Innes, T.C.; Bouwer, G. An improved bioassay for the detection of Bacillus thuringiensis beta-exotoxin. J. Invertebr. Pathol. 2009, 101, 137–139. [Google Scholar] [CrossRef]
- Palma, L.; Hernández-Rodríguez, C.S.; Maeztu, M.; Hernández-Martínez, P.; Ruiz de Escudero, I.; Escriche, B.; Muñoz, D.; van Rie, J.; Ferré, J.; Caballero, P. Vip3C, a novel class of vegetative insecticidal proteins from Bacillus thuringiensis. Appl. Environ. Microbiol. 2012, 78, 7163–7165. [Google Scholar] [CrossRef]
- Palma, L.; Ruiz de Escudero, I.; Maeztu, M.; Caballero, P.; Muñoz, D. Screening of vip genes from a Spanish Bacillus thuringiensis collection and characterization of two novel Vip3 proteins highly-toxic to five lepidopteran crop pests. Biol. Control 2013, 66, 141–149. [Google Scholar] [CrossRef]
- Tan, F.; Zhu, J.; Tang, J.; Tang, X.; Wang, S.; Zheng, A.; Li, P. Cloning and characterization of two novel crystal protein genes, cry54Aa1 and cry30Fa1, from Bacillus thuringiensis strain BtMC28. Curr. Microbiol. 2009, 58, 654–659. [Google Scholar] [CrossRef]
- Metzker, M.L. Sequencing technologies—The next generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef]
- Shendure, J.; Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 2008, 26, 1135–1145. [Google Scholar] [CrossRef]
- Iriarte, J.; Bel, Y.; Ferrandis, M.D.; Andrew, R.; Murillo, J.; Ferré, J.; Caballero, P. Environmental distribution and diversity of Bacillus thuringiensis in Spain. Syst. Appl. Microbiol. 1998, 21, 97–106. [Google Scholar] [CrossRef]
- Iriarte, J.; Porcar, M.; Lecadet, M.; Caballero, P. Isolation and characterization of Bacillus thuringiensis strains from aquatic environments in Spain. Curr. Microbiol. 2000, 40, 402–408. [Google Scholar] [CrossRef]
- Porcar, M.; Caballero, P. Molecular and insecticidal characterization of a Bacillus thuringiensis strain isolated during a natural epizootic. J. Appl. Microbiol. 2000, 89, 309–316. [Google Scholar] [CrossRef]
- Martínez, C.; Porcar, M.; López, A.; Ruiz de Escudero, I.; Pérez-Llarena, F.J.; Caballero, P. Characterization of a Bacillus thuringiensis strain with a broad spectrum of activity against lepidopteran insects. Entomol. Exp. Appl. 2004, 111, 71–77. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Ye, W.; Zhu, L.; Liu, Y.; Crickmore, N.; Peng, D.; Ruan, L.; Sun, M. Mining new crystal protein genes from Bacillus thuringiensis based on mixed plasmid-enriched genome sequencing and a computational pipeline. Appl. Environ. Microbiol. 2012, 78, 4795–4801. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genomics 2008, 9, 1–15. [Google Scholar] [CrossRef]
- Pruss, B.M.; Dietrich, R.; Nibler, B.; Martlbauer, E.; Scherer, S. The hemolytic enterotoxin HBL is broadly distributed among species of the Bacillus cereus group. Appl. Environ. Microbiol. 1999, 65, 5436–5442. [Google Scholar]
- Drummond, A.J.; Ashton, B.; Buxton, S.; Cheung, M.; Cooper, A.; Duran, C.; Field, M.; Heled, J.; Kearse, M.; Markowitz, S.; et al. Geneious Pro v6.1.7. Available online: http://www.geneious.com/ (accessed on 01 April 2014).
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Pence, R.J. The antimetabolite, imidazole as a pesticide. J. Econ. Entomol. 1963, 56, 1–7. [Google Scholar]
- Ross, D.C.; Brown, T.M. Inhibition of larval growth in Spodoptera frugiperda by sublethal dietary concentrations of insecticides. J. Agric. Food Chem. 1982, 30, 193–196. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Ferré, J.; van Rie, J. Biochemistry and genetics of insect resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 2002, 47, 501–533. [Google Scholar] [CrossRef]
- Sayyed, A.H.; Haward, R.; Herrero, S.; Ferré, J.; Wright, D.J. Genetic and biochemical approach for characterization of resistance to Bacillus thuringiensis toxin Cry1Ac in a field population of the diamondback moth, Plutella xylostella. Appl. Environ. Microbiol. 2000, 66, 1509–1516. [Google Scholar] [CrossRef]
- Oppert, B.; Kramer, K.J.; Beeman, R.W.; Johnson, D.; McGaughey, W.H. Proteinase-mediated insect resistance to Bacillus thuringiensis toxins. J. Biol. Chem. 1997, 272, 23473–23476. [Google Scholar] [CrossRef]
- Mardis, E.R. Next-generation DNA sequencing methods. Annu. Rev. Genomics Hum. Genet. 2008, 9, 387–402. [Google Scholar] [CrossRef]
- Zhou, X.; Ren, L.; Meng, Q.; Li, Y.; Yu, Y.; Yu, J. The next-generation sequencing technology and application. Protein Cell 2010, 1, 520–536. [Google Scholar] [CrossRef]
- Schuster, S.C. Next-generation sequencing transforms today’s biology. Nat. Methods 2008, 5, 16–18. [Google Scholar]
- Baltrus, D.A.; Nishimura, M.T.; Romanchuk, A.; Chang, J.H.; Mukhtar, M.S.; Cherkis, K.; Roach, J.; Grant, S.R.; Jones, C.D.; Dangl, J.L. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog. 2011, 7, e1002132. [Google Scholar] [CrossRef]
- Palma, L.; Universidad Pública de Navarra, Navarra, Spain. Unpublished work. 2014.
- Porcar, M.; Iriarte, J.; Dumanoir, V.C.; Ferrandis, M.D.; Lecadet, M.M.; Ferre, J.; Caballero, P. Identification and characterization of the new Bacillus thuringiensis serovars pirenaica (serotype H57) and iberica (serotype H59). J. Appl. Microbiol. 1999, 87, 640–648. [Google Scholar] [CrossRef]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef]
- Barton, M.D.; Barton, H.A. Scaffolder—Software for manual genome scaffolding. Source Code Biol. Med. 2012, 7. [Google Scholar] [CrossRef]
- Berry, C.; O’Neil, S.; Ben-Dov, E.; Jones, A.F.; Murphy, L.; Quail, M.A.; Holden, M.T.; Harris, D.; Zaritsky, A.; Parkhill, J. Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. israelensis. Appl. Environ. Microbiol. 2002, 68, 5082–5095. [Google Scholar] [CrossRef]
- De Maagd, R.A.; Bravo, A.; Berry, C.; Crickmore, N.; Schnepf, H.E. Structure, diversity, and evolution of protein toxins from spore-forming entomopathogenic bacteria. Annu. Rev. Genet. 2003, 37, 409–433. [Google Scholar] [CrossRef]
- McIver, K.S.; Myles, R.L. Two DNA-binding domains of Mga are required for virulence gene activation in the group A Streptococcus. Mol. Microbiol. 2002, 43, 1591–1601. [Google Scholar] [CrossRef]
- He, J.; Wang, J.P.; Yin, W.; Shao, X.H.; Zheng, H.J.; Li, M.S.; Zhao, Y.W.; Sun, M.; Wang, S.Y.; Yu, Z.N. Complete genome sequence of Bacillus thuringiensis subsp. chinensis strain CT-43. J. Bacteriol. 2011, 193, 3407–3408. [Google Scholar] [CrossRef]
- McClintock, J.; Stone, T.B.; Sjoblad, R.D. A comparative review of the mammalian toxicity of Bacillus thuringiensis-based pesticides. Pest Manag. Sci. 1995, 45, 95–105. [Google Scholar] [CrossRef]
- Siegel, J.P. The mammalian safety of Bacillus thuringiensis-based insecticides. J. Invertebr. Pathol. 2001, 77, 13–21. [Google Scholar] [CrossRef]
- Han, C.S.; Xie, G.; Challacombe, J.F.; Altherr, M.R.; Bhotika, S.S.; Brown, N.; Bruce, D.; Campbell, C.S.; Campbell, M.L.; Chen, J.; et al. Pathogenomic sequence analysis of Bacillus cereus and Bacillus thuringiensis isolates closely related to Bacillus anthracis. J. Bacteriol. 2006, 188, 3382–3390. [Google Scholar] [CrossRef]
- Hire, R.S.; Hadapad, A.B.; Dongre, T.K.; Kumar, V. Purification and characterization of mosquitocidal Bacillus sphaericus BinA protein. J. Invertebr. Pathol. 2009, 101, 106–111. [Google Scholar] [CrossRef]
- Ruiz de Escudero, I.; Estela, A.; Porcar, M.; Martínez, C.; Oguiza, J.A.; Escriche, B.; Ferré, J.; Caballero, P. Molecular and insecticidal characterization of a Cry1I protein toxic to insects of the families Noctuidae, Tortricidae, Plutellidae, and Chrysomelidae. Appl. Environ. Microbiol. 2006, 72, 4796–4804. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Palma, L.; Muñoz, D.; Berry, C.; Murillo, J.; Caballero, P. Draft Genome Sequences of Two Bacillus thuringiensis Strains and Characterization of a Putative 41.9-kDa Insecticidal Toxin. Toxins 2014, 6, 1490-1504. https://doi.org/10.3390/toxins6051490
Palma L, Muñoz D, Berry C, Murillo J, Caballero P. Draft Genome Sequences of Two Bacillus thuringiensis Strains and Characterization of a Putative 41.9-kDa Insecticidal Toxin. Toxins. 2014; 6(5):1490-1504. https://doi.org/10.3390/toxins6051490
Chicago/Turabian StylePalma, Leopoldo, Delia Muñoz, Colin Berry, Jesús Murillo, and Primitivo Caballero. 2014. "Draft Genome Sequences of Two Bacillus thuringiensis Strains and Characterization of a Putative 41.9-kDa Insecticidal Toxin" Toxins 6, no. 5: 1490-1504. https://doi.org/10.3390/toxins6051490
APA StylePalma, L., Muñoz, D., Berry, C., Murillo, J., & Caballero, P. (2014). Draft Genome Sequences of Two Bacillus thuringiensis Strains and Characterization of a Putative 41.9-kDa Insecticidal Toxin. Toxins, 6(5), 1490-1504. https://doi.org/10.3390/toxins6051490






