The Pore-Forming Haemolysins of Bacillus Cereus: A Review
Abstract
:1. Introduction
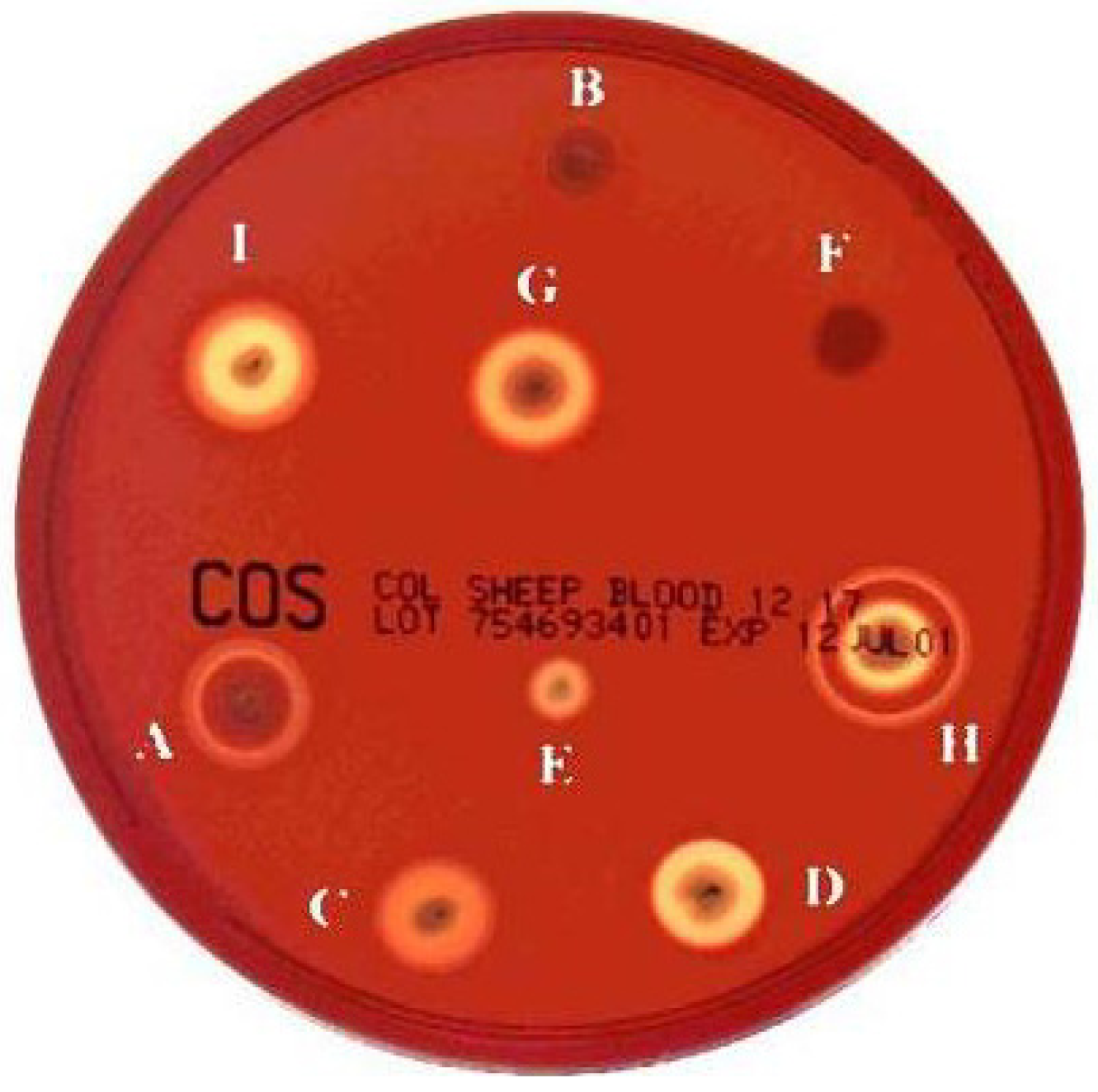
2. Haemolysin I (Cereolysin O)
2.1. Genomic and Structural Features

2.2. CLO in Virulence
2.3. Gene Regulation
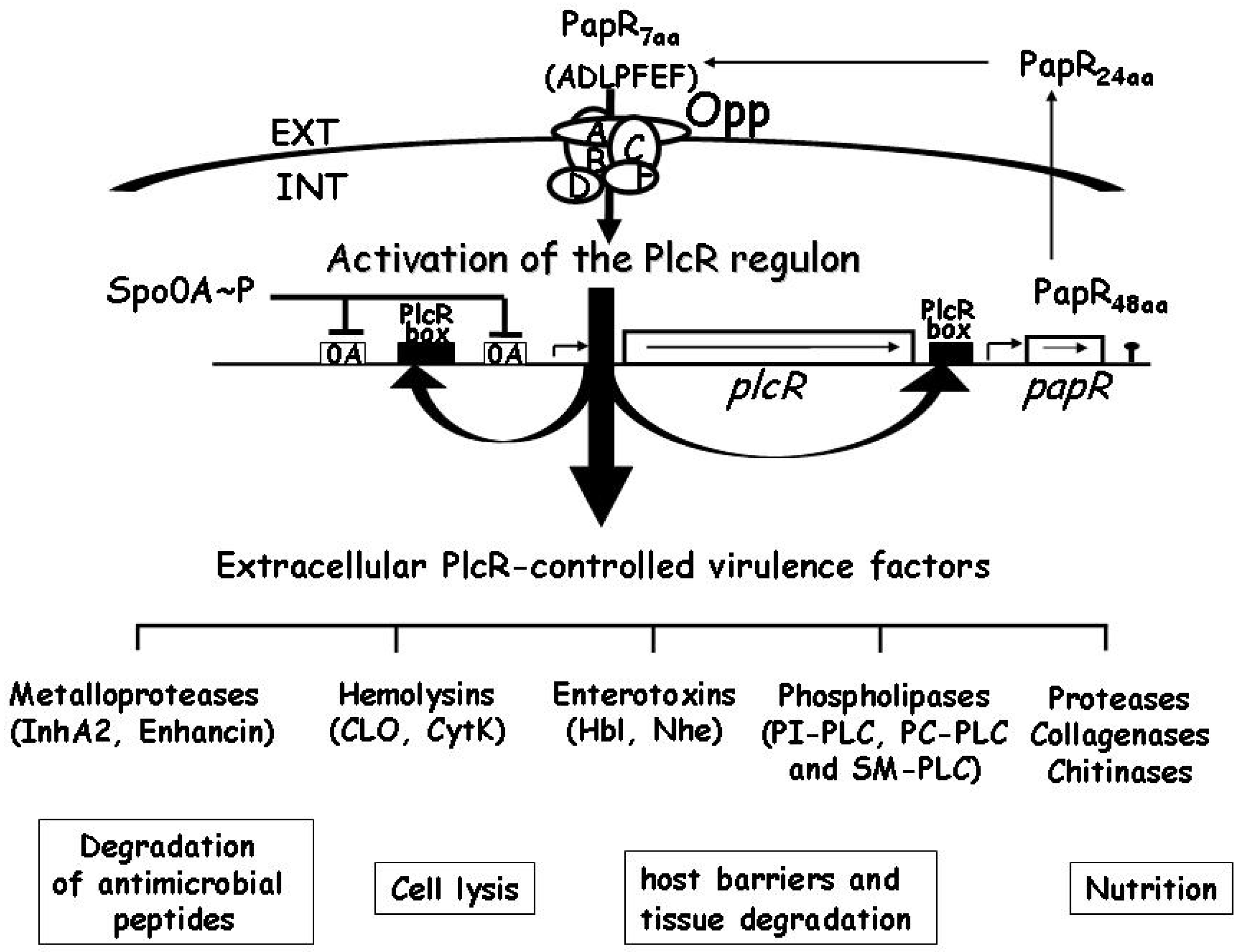
3. Haemolysin II
3.1. Genomic and Structural Features

3.2. Haemolysin II in Virulence
3.3. Regulation of Expression of the hlyII Gene

4. Haemolysin III
4.1. Genomic and Structural Features
4.2. HlyIII in Virulence
5. Haemolysin IV (Cytotoxin K)
5.1. Genomic and Structural Features
5.2. CytK in Virulence
5.3. Distribution of cytK Genes in B. cereus
5.4. Regulation of cytK Expression
6. Conclusions
Conflict of Interest
References
- Guinebretière, M.H.; Thompson, F.L.; Sorokin, A.; Normand, P.; Dawyndt, P.; Ehling-Schulz, M.; Svensson, B.; Sanchis, V.; Nguyen-The, C.; Heyndrickx, M.; et al. Ecological diversification in the Bacillus cereus Group. Environ. Microbiol. 2008, 10, 851–865. [Google Scholar] [CrossRef]
- Ivanova, N.; Sorokin, A.; Anderson, I.; Galleron, N.; Candelon, B.; Kapatral, V.; Bhattacharyya, A.; Reznik, G.; Mikhailova, N.; Lapidus, A.; et al. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 2003, 423, 87–91. [Google Scholar] [CrossRef]
- Rejasse, A.; Gilois, N.; Barbosa, I.; Huillet, E.; Bevilacqua, C.; Tran, S.; Ramarao, N.; Stenfors, A.L.P.; Sanchis, V. Temperature-dependent production of various PlcR-controlled virulence factors in Bacillus weihenstephanensis strain KBAB4. Appl. Environ. Microbiol. 2012, 78, 2553–2561. [Google Scholar] [CrossRef]
- Kolstø, A.B.; Tourasse, N.J.; Økstad, O.A. What sets Bacillus anthracis apart from other Bacillus species? Annu. Rev. Microbiol. 2009, 63, 451–476. [Google Scholar] [CrossRef]
- Anonymous. The community summary report on food-borne outbreaks in the European Union in 2007. EFSA J. 2009. [Google Scholar] [CrossRef]
- Hernaiz, C.; Picardo, A.; Alos, J.I.; Gomez-Garces, J.L. Nosocomial bacteremia and catheter infection by Bacillus cereus in an immunocompetent patient. Clin. Microbiol. Infect. 2003, 9, 973–975. [Google Scholar] [CrossRef]
- Kotiranta, A.; Lounatmaa, K.; Haapasalo, M. Epidemiology and pathogenesis of Bacillus cereus infections. Microbes Infect. 2000, 2, 189–198. [Google Scholar] [CrossRef]
- Lund, T.; de Buyser, M.L.; Granum, P.E. A new cytotoxin from Bacillus cereus that may cause necrotic enteritis. Mol. Microbiol. 2000, 38, 254–261. [Google Scholar] [CrossRef]
- Naranjo, M.; Denayer, S.; Botteldoorn, N.; Delbrassinne, L.; Veys, J.; Waegenaere, J.; Sirtaine, N.; Driesen, R.B.; Sipido, K.R.; Mahillon, J.; et al. Sudden death of a young adult associated with Bacillus cereus food poisoning. J. Clin. Microbiol. 2011, 49, 4379–4381. [Google Scholar] [CrossRef]
- Mahler, H.; Pasi, A.; Kramer, J.M.; Schulte, P.; Scoging, A.C.; Bär, W.; Krähenbühl, S. Fulminant liver failure in association with the emetic toxin of Bacillus cereus. N. Engl. J. Med. 1997, 336, 1142–1148. [Google Scholar] [CrossRef]
- Dierick, K.; van Coillie, E.; Swiecicka, I.; Meyfroidt, G.; Devlieger, H.; Meulemans, A.; Hoedemaekers, G.; Fourie, L.; Heyndrickx, M.; Mahillon, J. Fatal family outbreak of Bacillus cereus-associated food poisoning. J. Clin. Microbiol. 2005, 43, 4277–4279. [Google Scholar] [CrossRef]
- Ehling-Schulz, M.; Fricker, M.; Scherer, S. Bacillus cereus, the causative agent of an emetic type of food-borne illness. Mol. Nutr. Food Res. 2004, 48, 479–487. [Google Scholar] [CrossRef]
- Stenfors, A.L.P.; Fagerlund, A.; Granum, P.E. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 2008, 32, 579–606. [Google Scholar] [CrossRef]
- Ramarao, N. Bacillus cereus: Caractéristiques et pathogénicité. EMC Biol. Méd. 2012, 7, 1–11. [Google Scholar]
- Cadot, C.; Tran, S.L.; Vignaud, M.L.; de Buyser, M.L.; Kolstø, A.B.; Brisabois, A.; Nguyen-Thé, C.; Lereclus, D.; Guinebretière, M.H.; Ramarao, N. InhA1, NprA and HlyII as candidates to differentiate pathogenic from non-pathogenic Bacillus cereus strains. J. Clin. Microbiol. 2010, 48, 1358–1365. [Google Scholar] [CrossRef]
- Bottone, E.J. Bacillus cereus, a volatile human pathogen. Clin. Microbiol. Rev. 2010, 23, 382–398. [Google Scholar] [CrossRef]
- Callegan, M.C.; Kane, S.T.; Cochran, D.C.; Novosad, B.; Gilmore, M.S.; Gominet, M.; Lereclus, D. Bacillus endophthalmitis: Roles of bacterial toxins and motility during infection. Invest. Ophthalmol. Vis. Sci. 2005, 46, 3233–3238. [Google Scholar] [CrossRef]
- Gray, J.; George, R.H.; Durbin, G.M.; Ewer, A.K.; Hocking, M.D.; Morgan, M.E. An outbreak of Bacillus cereus respiratory tract infections on a neonatal unit due to contaminated ventilator circuits. J. Hosp. Infect. 1999, 41, 19–22. [Google Scholar] [CrossRef]
- Miller, J.M.; Hair, J.G.; Hebert, M.; Hebert, L.; Roberts, F.J., Jr; Weyant, R. Fulminating bacteremia and pneumonia due to Bacillus cereus. J. Clin. Microbiol. 1997, 35, 504–507. [Google Scholar]
- Bouillaut, L.; Ramarao, N.; Buisson, C.; Gilois, N.; Gohar, M.; Lereclus, D.; Nielsen-Leroux, C. FlhA influences Bacillus thuringiensis PlcR-regulated gene transcription, protein production, and virulence. Appl. Environ. Microbiol. 2005, 71, 8903–8910. [Google Scholar] [CrossRef]
- Tran, S.L.; Guillemet, E.; Gohar, M.; Lereclus, D.; Ramarao, N. CwpFM (EntFM) is a Bacillus cereus potential cell wall peptidase implicated in adhesion, biofilm formation and virulence. J. Bacteriol. 2010, 192, 2638–2642. [Google Scholar] [CrossRef]
- Gilois, N.; Ramarao, N.; Bouillaut, L.; Perchat, S.; Aymerich, S.; Nielsen-Leroux, C.; Lereclus, D. Growth-related variations in the Bacillus cereus secretome. Proteomics 2007, 7, 1719–1728. [Google Scholar] [CrossRef]
- Brillard, J.; Susanna, K.; Michaud, C.; Dargaignaratz, C.; Gohar, M.; Nielsen-Leroux, C.; Ramarao, N.; Kolstø, A.B.; Nguyen-the, C.; Lereclus, D.; Broussolle, V. The YvfTU two-component system is involved in plcR expression in Bacillus cereus. BMC Microbiol. 2008. [Google Scholar] [CrossRef]
- Ramarao, N.; Lereclus, D. Adhesion and cytotoxicity of Bacillus cereus and Bacillus thuringiensis to epithelial cells are FlhA and PlcR dependent, respectively. Microbes Infect. 2006, 8, 1483–1491. [Google Scholar] [CrossRef]
- Auger, S.; Ramarao, N.; Faille, C.; Fouet, A.; Aymerich, S.; Gohar, M. Biofilm formation and cell surface properties among pathogenic and non pathogenic strains of the Bacillus cereus group. Appl. Environ. Microbiol. 2009, 75, 6616–6618. [Google Scholar] [CrossRef]
- Guillemet, E.; Cadot, C.; Tran, S.L.; Guinebretière, M.H.; Lereclus, D.; Ramarao, N. The InhA metalloproteases of Bacillus cereus contribute concomitantly to virulence. J. Bacteriol. 2010, 192, 286–294. [Google Scholar] [CrossRef]
- Lindbäck, T.; Okstad, O.A.; Rishovd, A.L.; Kolstø, A.B. Insertional inactivation of hblC encoding the L2 component of Bacillus cereus ATCC 14579 haemolysin BL strongly reduces enterotoxigenic activity, but not the haemolytic activity against human erythrocytes. Microbiology 1999, 145, 3139–3146. [Google Scholar]
- Fagerlund, A.; Lindbäck, T.; Storset, A.K.; Granum, P.E.; Hardy, S.P. Bacillus cereus Nhe is a pore forming toxin with structural and functional properties similar to ClyA (HlyE, SheA) family of haemolysins, able to induce osmotic lysis in epithelia. Microbiology 2008, 154, 693–704. [Google Scholar] [CrossRef]
- Senesi, S.; Ghelardi, E. Production, secretion and biological activity of Bacillus cereus enterotoxins. Toxins 2010, 2, 1690–1703. [Google Scholar] [CrossRef]
- Gilmore, M.S.; Cruz-Rodz, A.L.; Leimeister-Wächter, M.; Kreft, J.; Goebel, W. A Bacillus cereus cytolitic determinant, cereolysin AB, which comprises the phospholipase C and sphingomyelinase genes: Nucleotide sequence and genetic linkage. J. Bacteriol. 1989, 171, 744–753. [Google Scholar]
- Vilas-Boas, G.; Sanchis, V.; Lereclus, D.; Lemos, M.V.; Bourguet, D. Genetic differentiation between sympatric populations of Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 2002, 68, 1414–1424. [Google Scholar] [CrossRef]
- Bernheimer, A.W.; Grushoff, P. Cereolysin: Production, purification and partial characterization. J. Gen. Microbiol. 1967, 46, 143–150. [Google Scholar] [CrossRef]
- Shannon, J.G.; Ross, C.L.; Koehler, T.M.; Rest, R.F. Characterization of anthrolysin O, the Bacillus anthracis cholesterol-dependent cytolysin. Infect. Immun. 2003, 71, 3183–3189. [Google Scholar] [CrossRef]
- Johnson, M.K.; Geoffroy, C.; Alouf, J.E. Binding of cholesterol by sulfhydryl-activated cytolysins. Infect. Immun. 1980, 27, 97–101. [Google Scholar]
- Bernheimer, A.W.; Grushoff, P. Extracellular hemolysins of aerobic sporogenic Bacilli. J. Bacteriol. 1967, 93, 1541–1543. [Google Scholar]
- Alouf, J.E.; Billington, S.J.; Jost, B.H. Repertoire and General Features of the Family of Cholesterol-Dependent Cytolysins. In The Comprehensive Sourcebook of Bacterial Protein Toxins, 3rd ed.; Alouf, J.E., Popoff, M.R., Eds.; Academic Press: London, UK, 2005; pp. 643–658. [Google Scholar]
- Palmer, M. The family of thiol-activated, cholesterol-binding cytolysins. Toxicon 2001, 39, 1681–1689. [Google Scholar] [CrossRef]
- Shatursky, O.; Heuck, A.P.; Shepard, L.A.; Rossjohn, J.; Parker, M.W.; Johnson, A.E.; Tweten, R.K. The mechanism of membrane insertion for a cholesterol-dependent cytolysin: A novel paradigm for pore-forming toxins. Cell 1999, 99, 293–299. [Google Scholar] [CrossRef]
- Jacobs, T.; Cima-Cabal, M.D.; Darji, A.; Méndez, F.J.; Vázquez, F.; Jacobs, A.A.; Shimada, Y.; Ohno-Iwashita, Y.; Weiss, S.; de los Toyos, J.R. The conserved undecapeptide shared by thiol-activated cytolysins is involved in membrane binding. FEBS Lett. 1999, 459, 463–466. [Google Scholar] [CrossRef]
- Rossjohn, J.; Feil, S.C.; McKinstry, W.J.; Tweten, R.K.; Parker, M.W. Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form. Cell 1997, 89, 685–692. [Google Scholar] [CrossRef]
- Polekhina, G.; Giddings, K.S.; Tweten, R.K.; Parker, M.W. Insights into the action of the superfamily of cholesterol-dependent cytolysins from studies of intermedilysin. Proc. Natl. Acad. Sci. USA 2005, 102, 600–605. [Google Scholar] [CrossRef]
- Bourdeau, R.W.; Malito, E.; Chenal, A.; Bishop, B.L.; Musch, M.W.; Villereal, M.L.; Chang, E.B.; Mosser, E.M.; Rest, R.F.; Tang, W.J. Cellular functions and X-ray structure of anthrolysin O, a cholesterol-dependent cytolysin secreted by Bacillus anthracis. J. Biol. Chem. 2009, 284, 14645–14656. [Google Scholar] [CrossRef]
- Soltani, C.E.; Hotze, E.M.; Johnson, A.E.; Tweten, R.K. Specific protein-membrane contacts are required for prepore and pore assembly by a cholesterol-dependent cytolysin. J. Biol. Chem. 2007, 282, 15709–15716. [Google Scholar]
- Dang, T.X.; Hotze, E.M.; Rouiller, I.; Tweten, R.K.; Wilson-Kubalek, E.M. Prepore to pore transition of a cholesterol-dependent cytolysin visualized by electron microscopy. J. Struct. Biol. 2005, 150, 100–108. [Google Scholar] [CrossRef]
- Vazquez-Boland, J.A.; Kuhn, M.; Berche, P.; Chakraborty, T.; Domínguez-Bernal, G.; Goebel, W.; González-Zorn, B.; Wehland, J.; Kreft, J. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 2001, 14, 584–640. [Google Scholar] [CrossRef]
- Mosser, E.M.; Rest, R.F. The Bacillus anthracis cholesterol-dependent cytolysin, Anthrolysin O, kills human neutrophils, monocytes and macrophages. BMC Microbiol. 2006. [Google Scholar] [CrossRef]
- Beecher, D.J.; Olsen, T.W.; Somers, E.B.; Wong, A.C. Evidence for contribution of tripartite hemolysin BL, phosphatidylcholine-preferring phospholipase C, and collagenase to virulence of Bacillus cereus endophthalmitis. Infect. Immun. 2000, 68, 5269–5276. [Google Scholar] [CrossRef]
- Agaisse, H.; Gominet, M.; Okstad, O.A.; Kolstø, A.B.; Lereclus, D. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 1999, 32, 1043–1053. [Google Scholar] [CrossRef]
- Gohar, M.; Økstad, O.A.; Gilois, N.; Sanchis, V.; Kolstø, A.B.; Lereclus, D. Two-dimensional electrophoresis analysis of the extracellular proteome of Bacillus cereus reveals the importance of the PlcR regulon. Proteomics 2002, 2, 784–791. [Google Scholar] [CrossRef]
- Lereclus, D.; Agaisse, H.; Gominet, M.; Salamitou, S.; Sanchis, V. Identification of a Bacillus thuringiensis gene that positively regulates transcription of the phosphatidylinositol-specific phospholipase C gene at the onset of the stationary phase. J. Bacteriol. 1996, 178, 2749–2756. [Google Scholar]
- Gominet, M.; Slamti, L.; Gilois, N.; Rose, M.; Lereclus, D. Oligopeptide permease is required for expression of the Bacillus thuringiensis plcR regulon and for virulence. Mol. Microbiol. 2001, 40, 963–975. [Google Scholar] [CrossRef]
- Bouillaut, L.; Perchat, S.; Arold, S.; Zorrilla, S.; Slamti, L.; Henry, C.; Gohar, M.; Declerck, N.; Lereclus, D. Molecular basis for group-specific activation of the virulence regulator PlcR by PapR heptapeptides. Nucleic Acids Res. 2008, 36, 3791–801. [Google Scholar] [CrossRef]
- Lereclus, D.; Agaisse, H. Toxin and Virulence Gene Expression in Bacillus thuringiensis. In Entomopathogenic Bacteria: From Laboratory to Field Application; Charles, J.F., Delécluse, A., Nielsen-Leroux, C., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2000; pp. 127–142. [Google Scholar]
- Slamti, L.; Lereclus, D. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J. 2002, 21, 4550–4559. [Google Scholar] [CrossRef]
- Gohar, M.; Faegri, K.; Perchat, S.; Ravnum, S.; Økstad, O.A.; Gominet, M.; Kolstø, A.B.; Lereclus, D. The PlcR virulence regulon of Bacillus cereus. PLoS One 2008, 3, e2793. [Google Scholar] [CrossRef]
- Coolbaugh, J.C.; Williams, R.P. Production and characterization of two hemolysins of Bacillus cereus. Can. J. Microbiol. 1978, 24, 1289–1295. [Google Scholar] [CrossRef]
- Sinev, M.A.; Budarina, Z.I.; Gavrilenko, I.V.; Tomashevskiĭ, A.I.; Kuzmin, N.P. Evidence of the existence of hemolysin II from Bacillus cereus: Cloning the genetic determinant of hemolysin II. Mol. Biol. 1993, 27, 1218–1229. [Google Scholar]
- Baida, G.; Budarina, Z.I.; Kuzmin, N.P.; Solonin, A.S. Complete nucleotide sequence and molecular characterization of hemolysin II gene from Bacillus cereus. FEMS Microbiol. Lett. 1999, 180, 7–14. [Google Scholar] [CrossRef]
- Andreeva, Z.; Nesterenko, V.F.; Yurkov, I.S.; Budarina, Z.I.; Sineva, E.V.; Solonin, A.S. Purification and cytotoxic properties of Bacillus cereus hemolysin II. Prot. Express. Purif. 2006, 47, 186–193. [Google Scholar] [CrossRef]
- Andreeva, Z.I.; Nesterenko, V.F.; Fomkina, M.G.; Ternovsky, V.I.; Suzina, N.E.; Bakulina, A.Y.; Solonin, A.S.; Sineva, E.V. The properties of Bacillus cereus hemolysin II pores depend on environmental conditions. Biochem. Biophys. Acta 2007, 1768, 253–263. [Google Scholar]
- Andreeva-Kovalevskaya, Z.I.; Solonin, A.S.; Sineva, E.V.; Ternovsky, V.I. Pore-forming proteins and adaptation of living organisms to environmental conditions. Biochemistry 2008, 73, 1473–1492. [Google Scholar]
- Miles, G.; Bayley, H.; Cheley, S. Properties of Bacillus cereus hemolysin II: A heptameric transmembrane pore. Protein Sci. 2006, 11, 1813–1824. [Google Scholar] [CrossRef]
- Tran, S.L.; Guillemet, E.; Ngo-Camus, M.; Clybouw, C.; Puhar, A.; Moris, A.; Gohar, M.; Lereclus, D.; Ramarao, N. Hemolysin II is a Bacillus cereus virulence factor that induces apoptosis of macrophages. Cell. Microbiol. 2011, 13, 92–108. [Google Scholar] [CrossRef]
- Tran, S.L.; Puhar, A.; Ngo-Camus, M.; Ramarao, N. Trypan blue dye enters viable cells incubated with the pore-forming toxin HlyII of Bacillus cereus. PLoS One 2011, 6, e22876. [Google Scholar]
- Valeva, A.; Hellmann, N.; Walev, I.; Strand, D.; Plate, M.; Boukhallouk, F.; Brack, A.; Hanada, K.; Decker, H.; Bhakdi, S. Evidence that clustered phosphocholine head groups serve as sites for binding and assembly of an oligomeric protein pore. J. Biol. Chem. 2006, 281, 26014–26021. [Google Scholar] [CrossRef]
- Liang, X.; Ji, Y. Involvment of alpha5beta1 integrin and TNF alpha in Staphylococcus aureus alpha toxin induced death of epithelial cells. Cell. Microbiol. 2007, 9, 1809–1821. [Google Scholar] [CrossRef]
- Nagahama, M.; Hayashhi, S.; Morimitsu, S.; Sakurai, J. Biological activities and pore formation of Clostridium perfringens beta toxin in HL 60 cells. J. Biol. Chem. 2003, 278, 36934–36941. [Google Scholar]
- Granum, P.E. Clostridium perfringens toxins involved in food poisoning. Int. J. Food. Microbiol. 1990, 10, 101–111. [Google Scholar] [CrossRef]
- Clair, G.; Roussi, S.; Armengaud, J.; Duport, C. Expanding the known repertoire of virulence factors produced by Bacillus cereus through early secretome profiling in three redox conditions. Mol. Cell. Proteomics 2010, 9, 1486–1498. [Google Scholar] [CrossRef]
- Hernandez, E.; Ramisse, F.; Ducoureau, J.P.; Cruel, T.; Cavallo, J.D. Bacillus thuringiensis subsp. konkukian (serotype H34) superinfection: case report and experimental evidence of pathogenicity in immunosuppressed mice. J. Clin. Microbiol. 1998, 36, 2138–2139. [Google Scholar]
- Ramarao, N.; Lereclus, D. The InhA1 metalloprotease allows spores of the B. cereus group to escape macrophages. Cell. Microbiol. 2005, 7, 1357–1364. [Google Scholar] [CrossRef]
- Sineva, E.; Andreeva-Kovalevskaya, Z.I.; Shadrin, A.M.; Gerasimov, Y.L.; Ternovsky, V.I.; Teplova, V.V.; Yurkova, T.V.; Solonin, A.S. Expression of Bacillus cereus hemolysin II in Bacillus subtilis renders the bacteria pathogenic for the crustacean Daphnia magna. FEMS Microbiol. Lett. 2009, 299, 110–119. [Google Scholar] [CrossRef]
- Budarina, Z.I.; Sinev, M.A.; Mayorov, S.G.; Tomashevski, A.Y.; Shmelev, I.V.; Kuzmin, N.P. Hemolysin II is more characteristic of Bacillus thuringiensis than Bacillus cereus. Arch. Microbiol. 1994, 161, 252–257. [Google Scholar]
- Budarina, Z.I.; Nikitin, D.V.; Zenkin, N.; Zakharova, M.; Semenova, E.; Shlyapnikov, M.G.; Rodikova, E.A.; Masyukova, S.; Ogarkov, O.; Baida, G.E.; et al. A new Bacillus cereus DNA-binding protein, HlyIIR, negatively regulates expression of B. cereus haemolysin II. Microbiology 2004, 150, 3691–3701. [Google Scholar] [CrossRef]
- Guillemet, E.; Tran, S.L.; Cadot, C.; Rognan, D.; Lereclus, D.; Ramarao, N. Glucose 6P binds and activates HlyIIR to repress Bacillus cereus haemolysin hlyII gene expression. PLoS One 2013, 8, e55085. [Google Scholar]
- Sineva, E.; Shadrin, A.; Rodikova, E.A.; Andreeva-Kovalevskaya, Z.I.; Protsenko, A.S.; Mayorov, S.G.; Galaktionova, D.Y.; Magelky, E.; Solonin, A.S. Iron regulates expression of Bacillus cereus hemolysin II via global regulator Fur. J. Bacteriol. 2012, 194, 3327–3335. [Google Scholar] [CrossRef]
- Tran, S.; Guillemet, E.; Lereclus, D.; Ramarao, N. Iron regulates Bacillus thuringiensis haemolysin hlyII gene expression during insect infection. J. Invert. Pathol. 2013, 113, 205–208. [Google Scholar] [CrossRef]
- Rodikova, E.A.; Kovalevskiy, O.V.; Mayorov, S.G.; Budarina, Z.I.; Marchenkov, V.V.; Melnik, B.S.; Leech, A.P.; Nikitin, D.V.; Shlyapnikov, M.G.; Solonin, A.S. Two HlyIIR dimers bind to a long perfect inverted repeat in the operator of the hemolysin II gene from Bacillus cereus. FEBS Lett. 2007, 581, 1190–1196. [Google Scholar] [CrossRef]
- Kovalevskiy, O.V.; Lebedev, A.A.; Surin, A.K.; Solonin, A.S.; Antson, A.A. Crystal structure of Bacillus cereus HlyIIR, a transcriptional regulator of the gene for pore-forming toxin hemolysin II. J. Mol. Biol. 2007, 365, 825–834. [Google Scholar] [CrossRef]
- Deutscher, J. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2008, 11, 87–93. [Google Scholar] [CrossRef]
- Harvie, D.R.; Vílchez, S.; Steggles, J.R.; Ellar, D.J. Bacillus cereus Fur regulates iron metabolism and is required for full virulence. Microbiology 2005, 151, 569–577. [Google Scholar] [CrossRef]
- Ratledge, C.; Dover, L.G. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 2000, 54, 881–941. [Google Scholar] [CrossRef]
- Weinberg, E.D. Iron availability and infection. Biochim. Biophys. Acta 2009, 1790, 600–605. [Google Scholar]
- Baida, G.E.; Kuzmin, N.P. Cloning and primary structure of a new hemolysin gene from Bacillus cereus. Biochim. Biophys. Acta 1995, 1264, 151–154. [Google Scholar]
- Hofmann, K.; Stoffel, W. TMbase—A database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 1993, 374, 166. [Google Scholar]
- Baida, G.E.; Kuzmin, N.P. Mechanism of action of hemolysin III from Bacillus cereus. Biochim. Biophys. Acta 1996, 1284, 122–124. [Google Scholar] [CrossRef]
- Beecher, D.J.; Wong, A.C. Cooperative, synergistic and antagonistic haemolytic interactions between haemolysin BL, phosphatidylcholine phospholipase C and sphingomyelinase from Bacillus cereus. Microbiology 2000, 146, 3033–3039. [Google Scholar]
- Guinebretiere, M.H.; Auger, S.; Galleron, N.; Contzen, M.; de Sarrau, B.; Debuyser, M.L.; Lamberet, G.; Fagerlund, A.; Granum, P.E.; Lereclus, D.; et al. Bacillus cytotoxicus sp. nov. is a novel thermotolerant species of the Bacillus cereus group occasionally associated with food poisoning. Int. J. Syst. Evol. Microbiol. 2013, 63, 31–40. [Google Scholar] [CrossRef]
- Song, L.H.; Hobaugh, M.R.; Shustak, C.; Cheley, S.; Bayley, H.; Gouaux, J.E. Structure of staphylococcal alpha hemolysin, a heptameric transmembrane pore. Science 1996, 274, 1859–1866. [Google Scholar] [CrossRef]
- Menestrina, G.; Serra, M.D.; Prevost, G. Mode of action of beta-barrel pore-forming toxins of the staphylococcal alpha-hemolysin family. Toxicon 2001, 39, 1661–1672. [Google Scholar] [CrossRef]
- Fagerlund, A.; Ween, O.; Lund, T.; Hardy, S.P.; Granum, P.E. Genetic and functional analysis of the cytK family genes in Bacillus cereus. Microbiology 2004, 150, 2689–2697. [Google Scholar] [CrossRef]
- Guinebretiere, M.H.; Fagerlund, A.; Granum, P.E.; Nguyen-The, C. Rapid discrimination of cytK-1 and cytK-2 genes in Bacillus cereus strains by a novel duplex PCR system. FEMS Microbiol. Lett. 2006, 259, 74–80. [Google Scholar] [CrossRef]
- Fagerlund, A.; Lindback, T.; Granum, P.E. Bacillus cereus cytotoxins Hbl, Nhe and CytK are secreted via the Sec translocation pathway. BMC Microbiol. 2010. [Google Scholar] [CrossRef]
- Hardy, S.P.; Lund, T.; Granum, P.E. CytK toxin of Bacillus cereus forms pores in planar lipid bilayers and is cytotoxic to intestinal epithelia. FEMS Microbiol. Lett. 2001, 197, 47–51. [Google Scholar] [CrossRef]
- Ceuppens, S.; Rajkovic, A.; Heyndrickx, M.; Tsilia, V.; van de Wiele, T.; Boon, N.; Uyttendaele, M. Regulation of toxin production by Bacillus cereus and its food safety implications. Crit. Rev. Microbiol. 2011, 37, 188–213. [Google Scholar] [CrossRef]
- Kamar, R.; Gohar, M.; Jéhanno, I.; Réjasse, A.; Kallassy, M.; Lereclus, D.; Sanchis, V.; Ramarao, N. Pathogenic potential of B. cereus strains as revealed by phenotypic analysis. J. Clin. Microbiol. 2013, 51, 320–323. [Google Scholar] [CrossRef]
- Guinebretière, M.H.; Broussolle, V.; Nguyen-The, C. Enterotoxigenic profiles of food-poisoning and food-borne Bacillus cereus strains. J. Clin. Microbiol. 2002, 40, 3053–3056. [Google Scholar] [CrossRef]
- Wijnands, L.M.; Dufrenne, J.B.; Rombouts, F.M.; in ’t Veld, P.H.; van Leusden, F.M. Prevalence of potentially pathogenic Bacillus cereus in food commodities in The Netherlands. J. Food Prot. 2006, 69, 2587–2594. [Google Scholar]
- Ngamwongsatit, P.; Buasri, W.; Pianariyanon, P.; Pulsrikarn, C.; Ohba, M.; Assavanig, A.; Panbangred, W. Broad distribution of enterotoxin genes (hblCDA, nheABC, cytK, and entFM) among Bacillus thuringiensis and Bacillus cereus as shown by novel primers. Int. J. Food Microbiol. 2008, 121, 352–356. [Google Scholar] [CrossRef]
- Guinebretiere, M.H.; Velge, P.; Couvert, O.; Carlin, F.; Debuyser, M.L.; Nguyen-The, C. Ability of Bacillus cereus group strains to cause food poisoning varies according to phylogenetic affiliation (groups I to VII) rather than species affiliation. J. Clin. Microbiol. 2010, 48, 3388–3391. [Google Scholar]
- Brillard, J.; Lereclus, D. Comparison of cytotoxin cytK promoters from Bacillus cereus strain ATCC 14579 and from a B. cereus food-poisoning strain. Microbiology 2004, 150, 2699–2705. [Google Scholar] [CrossRef]
- Fagerlund, A.; Brillard, J.; Fürst, R.; Guinebretière, M.H.; Granum, P.E. Toxin production in a rare and genetically remote cluster of strains of the Bacillus cereus group. BMC Microbiol. 2007. [Google Scholar] [CrossRef]
- Ceuppens, S.; Timmery, S.; Mahillon, J.; Uyttendaele, M.; Boon, N. Small Bacillus cereus ATCC 14579 subpopulations are responsible for cytotoxin K production. J. Appl. Microbiol. 2013, 114, 899–906. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ramarao, N.; Sanchis, V. The Pore-Forming Haemolysins of Bacillus Cereus: A Review. Toxins 2013, 5, 1119-1139. https://doi.org/10.3390/toxins5061119
Ramarao N, Sanchis V. The Pore-Forming Haemolysins of Bacillus Cereus: A Review. Toxins. 2013; 5(6):1119-1139. https://doi.org/10.3390/toxins5061119
Chicago/Turabian StyleRamarao, Nalini, and Vincent Sanchis. 2013. "The Pore-Forming Haemolysins of Bacillus Cereus: A Review" Toxins 5, no. 6: 1119-1139. https://doi.org/10.3390/toxins5061119



