LRP1-Mediated Endocytosis May Be the Main Reason for the Difference in Cytotoxicity of Curcin and Curcin C on U2OS Osteosarcoma Cells
Abstract
:1. Introduction
2. Results
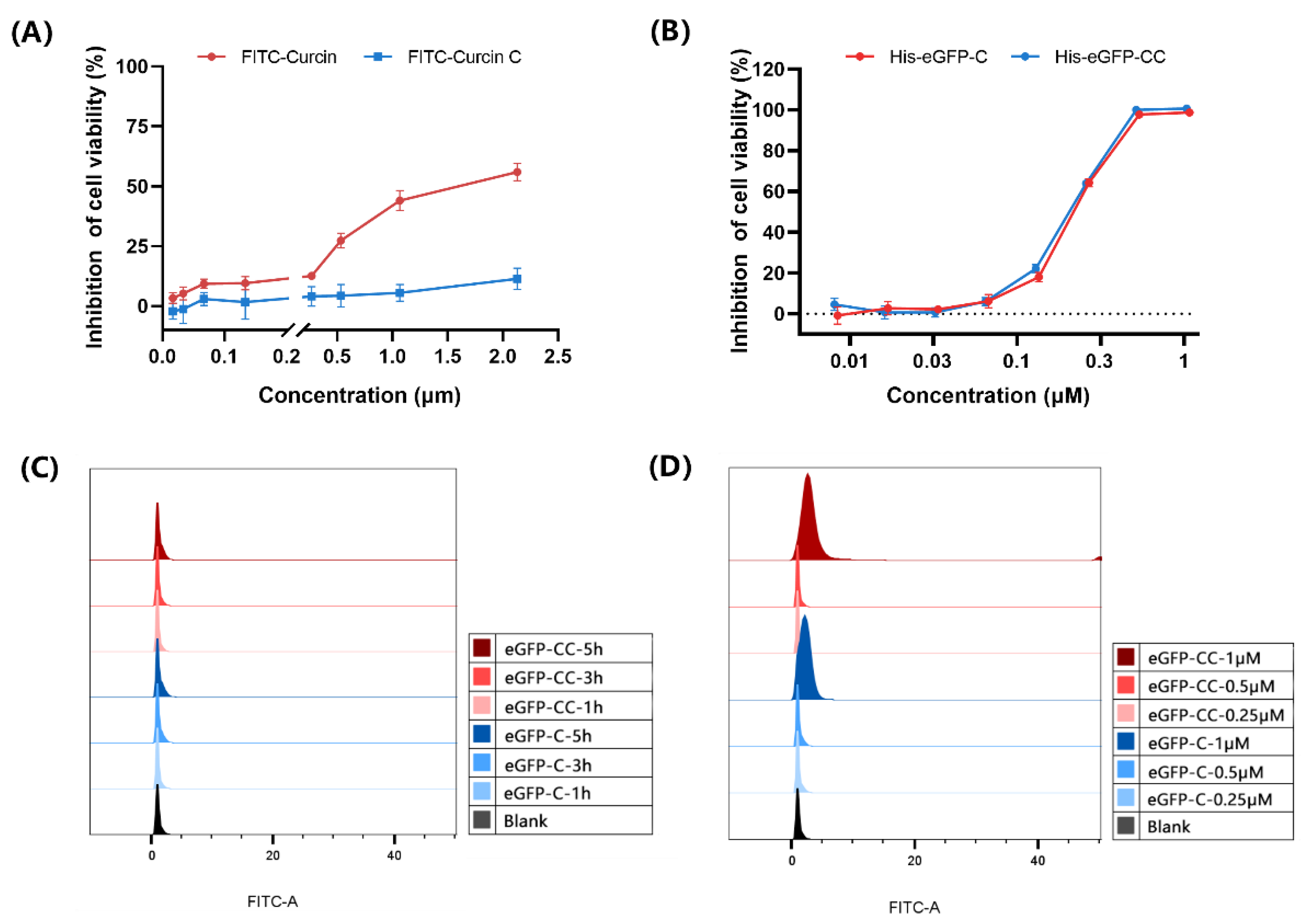
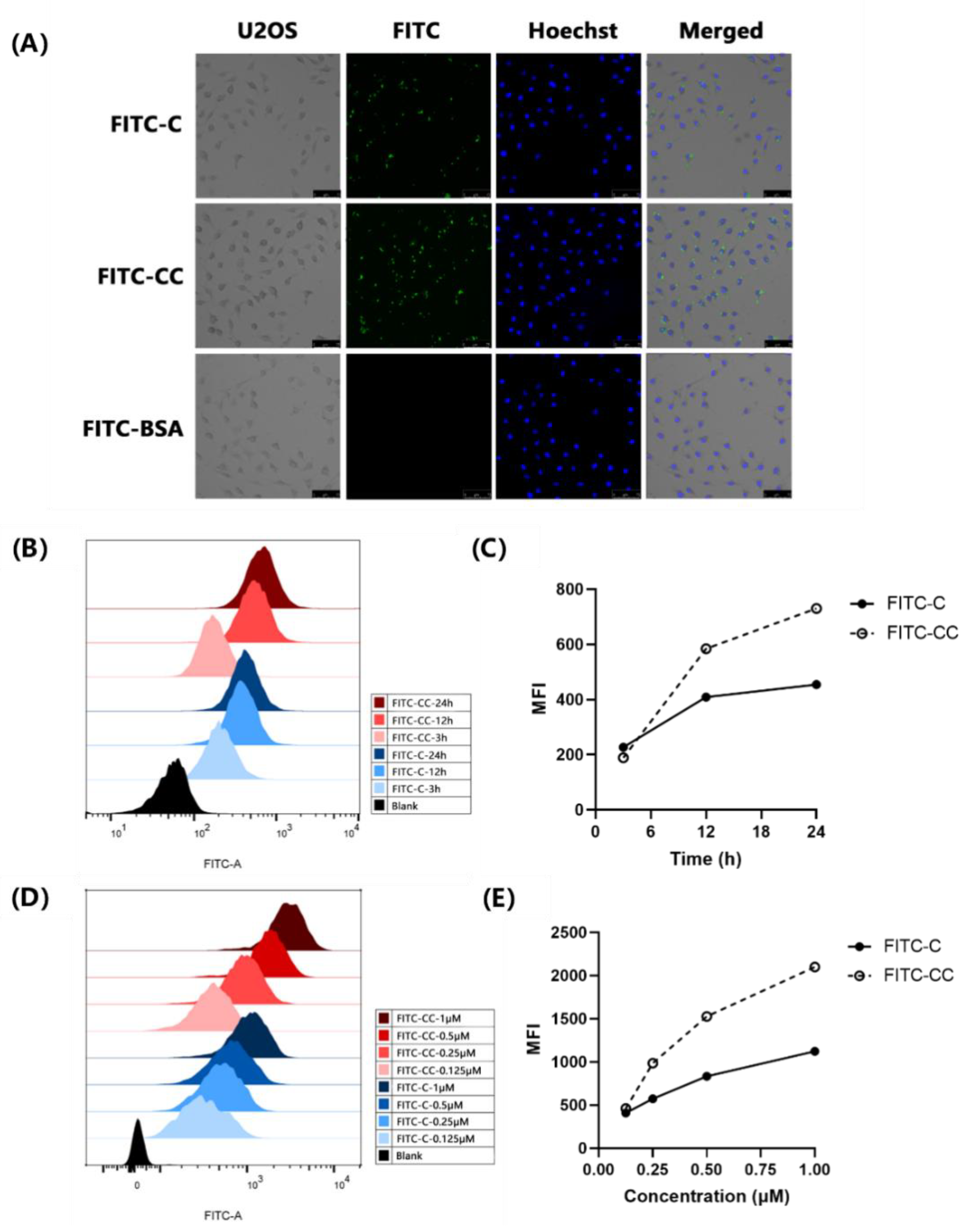
2.1. FITC-Curcin C Entered U2OS Cells More Efficiently Than FITC-Curcin
2.1.1. Comparison of Three Methods for Labeling Curcin and Curcin C
2.1.2. FITC-Curcin C Entered U2OS Cells More Efficiently Than FITC-Curcin
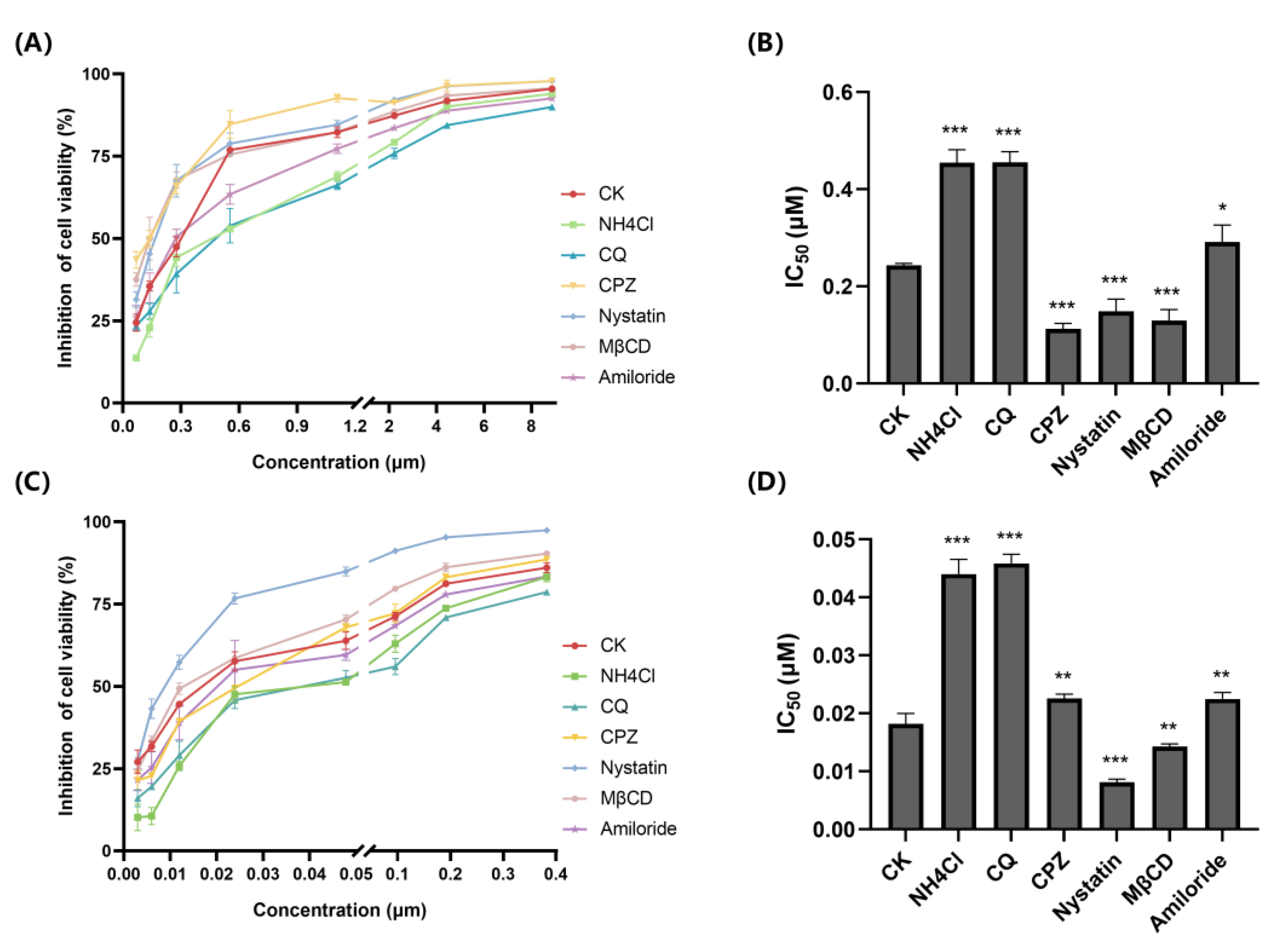
2.2. FITC-Curcin and FITC-Curcin C Have Different Entry Pathways
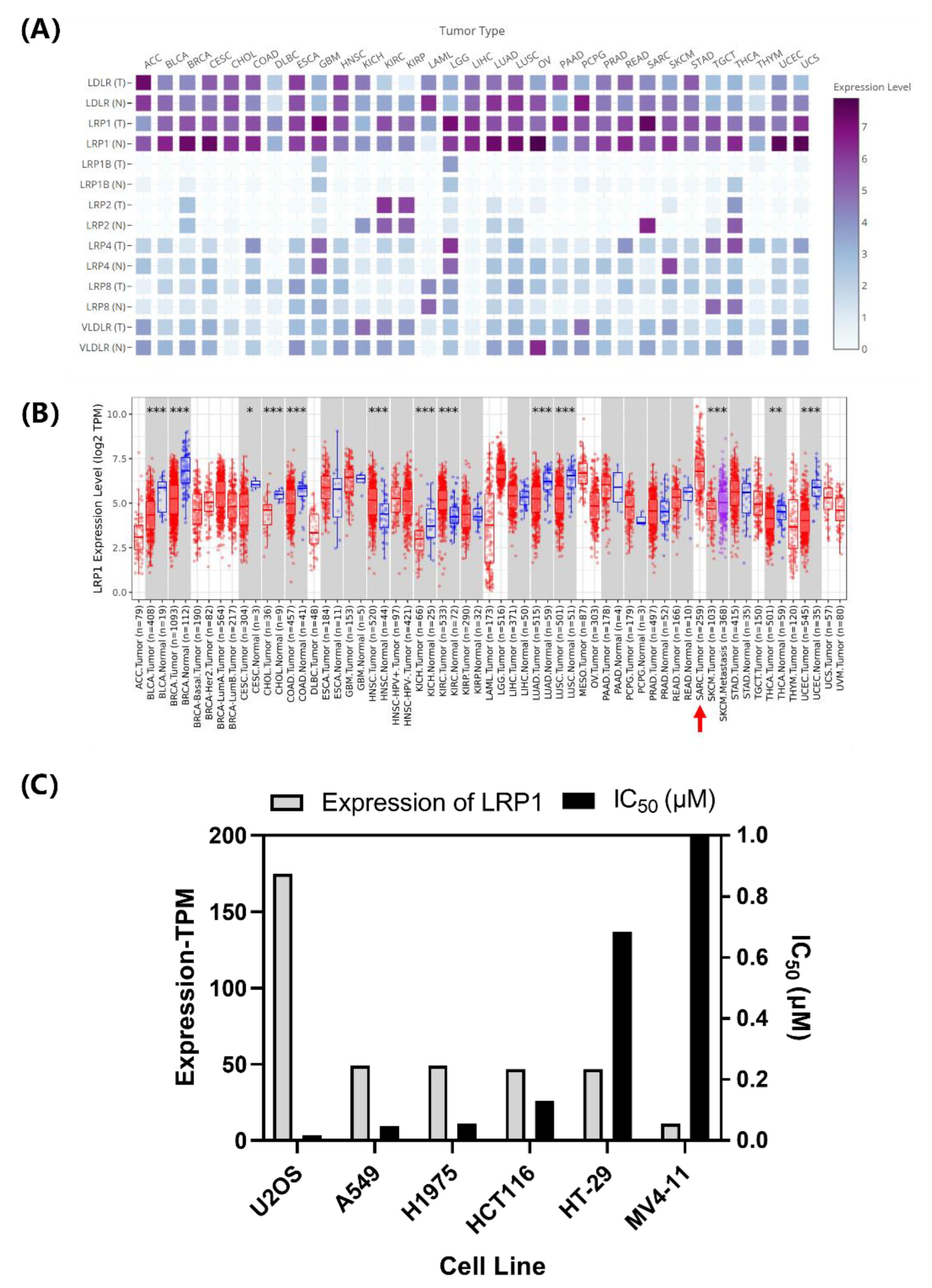
2.3. LRP1 Is One of Curcin C Endocytic Receptors
2.3.1. Correlation Analysis of LRP1 Abundance with Curcin and Curcin C Cytotoxicities
2.3.2. LRP1 Gene Silencing Partially Reduces the Cytotoxicity of Curcin C to U2OS Cells
2.3.3. Silencing LRP1 Partially Inhibits the Entry of FITC-Curcin C into U2OS Cells
3. Discussion
4. Materials and Methods
4.1. Materials and Cell Lines
4.2. FITC Labeling of Curcin, Curcin C, and BSA
4.3. Induced Expression of His-eGFP-Curcin and His-eGFP-Curcin C in Prokaryotes
4.4. Fluorescent Microscopy Observation of FITC-Curcin/Curcin C and His-eGFP-Curcin/Curcin C Entry into Cells
4.5. Flow Cytometry of FITC-Curcin/Curcin C and His-eGFP-Curcin/Curcin C in Cells
4.6. U2OS Cell Endocytic Inhibitors Pretreatment
4.7. CCK8 Assay for Cell Viability Characterization
4.8. LRP1 Gene Silencing in U2OS Cells by siRNA
4.9. Western Blotting for LRP1 Silencing Efficiency Characterization in U2OS Cells
4.10. Data Analysis and Processing
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lindsey, B.A.; Markel, J.E.; Kleinerman, E.S. Osteosarcoma Overview. Rheumatol. Ther. 2017, 4, 25–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrison, D.J.; Geller, D.S.; Gill, J.D.; Lewis, V.O.; Gorlick, R. Current and future therapeutic approaches for osteosarcoma. Expert Rev. Anti-Infect. Ther. 2017, 18, 39. [Google Scholar] [CrossRef] [PubMed]
- Saraf, A.J.; Fenger, J.M.; Roberts, R.D. Osteosarcoma: Accelerating Progress Makes for a Hopeful Future. Front. Oncol. 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crommelin, D.; Storm, G.; Verrijk, R.; Leede, L.D.; Jiskoot, W.; Hennink, W.E. Shifting paradigms: Biopharmaceuticals versus low molecular weight drugs. Int. J. Pharm. 2003, 266, 3–16. [Google Scholar] [CrossRef]
- Milletti, F. Cell-penetrating peptides: Classes, origin, and current landscape. Drug Discov. Today 2012, 17, 850–860. [Google Scholar]
- Barbier, J.; Gillet, D. Ribosome Inactivating Proteins: From Plant Defense to Treatments against Human Misuse or Diseases. Toxins 2018, 10, 160. [Google Scholar] [CrossRef] [Green Version]
- Rust, A.; Partridge, L.J.; Davletov, B.; Hautbergue, G.M. The Use of Plant-Derived Ribosome Inactivating Proteins in Immunotoxin Development: Past, Present and Future Generations. Toxins 2017, 9, 344. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.Y.; Tserng, K.Y.; Chen, C.C.; Lin, L.T.; Tung, T.C. Abrin and ricin: New anti-tumour substances. Nature 1970, 227, 292–293. [Google Scholar]
- Sha, O.; Niu, J.; Ng, T.B.; Cho, E.Y.-P. Anti-tumor action of trichosanthin, a type 1 ribosome-inactivating protein, employed in traditional Chinese medicine: A mini review. Cancer Chemother. Pharmacol. 2013, 71, 1387–1393. [Google Scholar] [CrossRef] [Green Version]
- Lee-Huang, S.; Huang, P.L.; Sun, Y.; Chen, H.C.; Kung, H.F.; Huang, P.L.; Murphy, W.J. Inhibition of MDA-MB-231 human breast tumor xenografts and HER2 expression by anti-tumor agents GAP31 and MAP30. Anticancer Res. 2000, 20, 653–659. [Google Scholar]
- Guo, J.L.; Cheng, Y.L.; Qiu, Y.; Shen, C.H.; Yi, B.; Peng, C. Purification and characterization of a novel type i ribosome inactivating protein, pachyerosin, from Pachyrhizus erosus seeds, and preparation of its immunotoxin against human hepatoma cells. Planta Med. 2014, 80, 896–901. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; He, L.; Meng, Y.; Li, G.; Li, L.; Meng, Y. A-MMC and MAP30, two ribosome-inactivating proteins extracted from Momordica charantia, induce cell cycle arrest and apoptosis in A549 human lung carcinoma cells. Mol. Med. Rep. 2015, 11, 3553–3558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Yang, Q.; Li, C.; Ding, M.; Lv, X.; Tao, C.; Yu, H.; Chen, F.; Xu, Y. Curcin C, a novel type I ribosome-inactivating protein from the post-germinating cotyledons of Jatropha curcas. Amino Acids 2017, 49, 1619–1631. [Google Scholar] [CrossRef] [PubMed]
- Conese, M.; Cavallaro, U.; Sidenius, N.; Olson, D.; Soria, M.R.; Blasi, F. PMA-induced down-regulation of the receptor for α2-macroglobulin in human U937 cells. FEBS Lett. 1995, 358, 73–78. [Google Scholar] [CrossRef] [Green Version]
- Vago, R.; Marsden, C.J.; Lord, J.M.; Ippoliti, R.; Flavell, D.J.; Flavell, S.U.; Ceriotti, A.; Fabbrini, M.S. Saporin and ricin A chain follow different intracellular routes to enter the cytosol of intoxicated cells. Febs J. 2010, 272, 4983–4995. [Google Scholar]
- Jiao, Y.Z.; Liu, W.Y. Low-density lipoprotein receptor-related protein 1 is an essential receptor for trichosanthin in 2 choriocarcinoma cell lines. Biochem. Biophys. Res. Commun. 2010, 391, 1579–1584. [Google Scholar] [CrossRef]
- Fabbrini, M.S.; Carpani, D.; Bello-Rivero, I.; Soria, M.R. The amino-terminal fragment of human urokinase directs a recombinant chimeric toxin to target cells: Internalization is toxin mediated. Faseb J. 1997, 11, 1169–1176. [Google Scholar]
- Fabbrini, M.S.; Rappocciolo, E.; Carpani, D.C.; Solinas, M.; Valsasina, B.; Breme, U.; Cavallaro, U.; Nykjaer, A.; Rovida, E.; Legname, G. Characterization of a saporin isoform with lower ribosome-inhibiting activity. Biochem. J. 1997, 322 Pt 3, 719–727. [Google Scholar] [CrossRef] [Green Version]
- Bolognesi, A.; Polito, L.; Scicchitano, V.; Orrico, C.; Pasquinelli, G.; Musiani, S.; Santi, S.; Riccio, M.; Bortolotti, M.; Battelli, M.G. Endocytosis and intracellular localisation of type 1 ribosome-inactivating protein saporin-s6. J. Biol. Regul. Homeost. Agents 2012, 26, 97–109. [Google Scholar]
- Schnaible, V.; Przybylski, M. Identification of fluorescein-5′-isothiocyanate-modification sites in proteins by electrospray-ionization mass spectrometry. Bioconjugate Chem. 1999, 10, 861–866. [Google Scholar]
- Lin, S.H.; Faller, L.D. Preparation of Na, K-ATPase specifically modified on the anti-fluorescein antibody-inaccessible site by fluorescein 5′-isothiocyanate. Anal. Biochem. 2000, 287, 303–312. [Google Scholar] [PubMed]
- Shi, B.-J.; Liu, C.-C.; Zhou, J.; Wang, S.-Q.; Gao, Z.-C.; Zhang, X.-M.; Zhou, B.; Chen, P.-Y. Entry of Classical Swine Fever Virus into PK-15 Cells via a pH-, Dynamin-, and Cholesterol-Dependent, Clathrin-Mediated Endocytic Pathway That Requires Rab5 and Rab7. J. Virol. 2016, 90, 9194–9208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawa, M.; Takasaki, T.; Yamada, K.-I.; Kurane, I.; Akatsuka, T. Interference in Japanese encephalitis virus infection of Vero cells by a cationic amphiphilic drug, chlorpromazine. J. Gen. Virol. 2003, 84 Pt 7, 1737–1741. [Google Scholar] [PubMed]
- Mayor, S.; Parton, R.G.; Donaldson, J.G. Clathrin-Independent Pathways of Endocytosis. Cold Spring Harb. Perspect. Biol. 2014, 6, a016758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Zhao, K.; Lan, Y.; Lv, X.; Hu, S.; Guan, J.; Lu, H.; Zhang, J.; Shi, J.; Yang, Y.; et al. Porcine Hemagglutinating Encephalomyelitis Virus Enters Neuro-2a Cells via Clathrin-Mediated Endocytosis in a Rab5-, Cholesterol-, and pH-Dependent Manner. J. Virol. 2017, 91, e01083-17. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.M.; Zhuang, X.Y.; Deng, Z.B.; Jiang, H.; Mu, J.Y.; Wang, Q.L.; Xiang, X.Y.; Guo, H.X.; Zhang, L.F.; Dryden, G.; et al. Targeted Drug Delivery to Intestinal Macrophages by Bioactive Nanovesicles Released from Grapefruit. Mol. Ther. 2014, 22, 522–534. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Shi, A.B. Endocytic recycling pathways and the regulatory mechanisms. Yi Chuan Hered. 2019, 41, 451–468. [Google Scholar]
- Maria, F.; Miku, K.; Ikuhiko, N.; Riccardo, V. Plant Ribosome-Inactivating Proteins: Progesses, Challenges and Biotechnological Applications (and a Few Digressions). Toxins 2017, 9, 314. [Google Scholar]
- Tang, N.; Chan, W.L.; Ke, Y.B.; Mak, M.; Lai, F.M.; Tam, S.C. Acute Renal Failure and Proximal Tubule Lesions after Trichosanthin Injection in Rats. Exp. Mol. Pathol. 1997, 64, 78–89. [Google Scholar] [CrossRef]
- Wang, F.; Wu, P.; Qin, S.; Deng, Y.; Han, P.; Li, X.; Fan, C.; Xu, Y. Curcin C inhibit osteosarcoma cell line U2OS proliferation by ROS induced apoptosis, autophagy and cell cycle arrest through activating JNK signal pathway. Int. J. Biol. Macromol. 2022, 195, 433–439. [Google Scholar] [CrossRef]
- Chan, W.L.; Shaw, P.C.; Tam, S.C.; Jacobsen, C.; Gliemann, J.; Nielsen, M.S. Trichosanthin Interacts with and Enters Cells via LDL Receptor Family Members. Biochem. Biophys. Res. Commun. 2000, 270, 453–457. [Google Scholar] [PubMed]
- Sandvig, K.; Bo, V.D. Transport of protein toxins into cells: Pathways used by ricin, cholera toxin and Shiga toxin. FEBS Lett. 2002, 529, 49–53. [Google Scholar] [CrossRef]
- Sandvig, K.; Deurs, B.V. Entry of ricin and Shiga toxin into cells: Molecular mechanisms and medical perspectives. Embo J. 2000, 19, 5943–5950. [Google Scholar] [PubMed] [Green Version]
- Sandvig, K.; Kavaliauskiene, S.; Skotland, T. The Protein Toxins Ricin and Shiga Toxin as Tools to Explore Cellular Mechanisms of Internalization and Intracellular Transport. Toxins 2021, 13, 377. [Google Scholar] [CrossRef] [PubMed]
- Roberts, L.M.; Lord, J.M. Ribosome-Inactivating Proteins: Entry into Mammalian Cells and Intracellular Routing. Mini Rev. Med. Chem. 2004, 4, 505–512. [Google Scholar] [CrossRef]
- Sandvig, K.; Torgersen, M.L.; Engedal, N.; Skotland, T.; Iversen, T.G. Protein toxins from plants and bacteria: Probes for intracellular transport and tools in medicine. Febs Lett. 2010, 584, 2626–2634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seaman, M.; Peden, A.A. Ricin Toxin Hits a Retrograde Roadblock. Cell 2010, 141, 222–224. [Google Scholar] [CrossRef] [Green Version]
- Römer, W.; Berland, L.; Chambon, V.; Gaus, K.; Windschiegl, B.; Tenza, D.; Aly, M.R.; Fraisier, V.; Florent, J.C.; Perrais, D.; et al. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature 2007, 450, 670–675. [Google Scholar] [CrossRef]
- Xia, X.F.; Sui, S.F. The membrane insertion of trichosanthin is membrane-surface-pH dependent. Biochem. J. 2000, 349 Pt 3, 835. [Google Scholar]
- Al-Bari, M.A.A. Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol. Res. Perspect. 2017, 5, e00293. [Google Scholar]
- Rapak, A.; Falnes, P.O.; Olsnes, S. Retrograde transport of mutant ricin to the endoplasmic reticulum with subsequent translocation to cytosol. Proc. Natl. Acad. Sci. USA 1997, 94, 3783–3788. [Google Scholar] [PubMed]
- García-Fernández, P.; Üçeyler, N.; Sommer, C. From the low-density lipoprotein receptor-related protein 1 to neuropathic pain: A potentially novel target. Pain Rep. 2021, 6, e898. [Google Scholar] [PubMed]
- Wang, L.; Shen, F.; Zhang, M.; He, Q.; Zhao, H.; Yu, X.; Yang, S.; Liu, Y.; Deng, N.; Zheng, J.; et al. Cytotoxicity mechanism of α-MMC in normal liver cells through LRP1 mediated endocytosis and JNK activation. Toxicology 2016, 357–358, 33–43. [Google Scholar] [CrossRef] [PubMed]

| Protein | Curcin | Curcin C | FITC-C | FITC-CC | His-eGFP-C | His-eGFP-CC |
|---|---|---|---|---|---|---|
| IC50 (nM) | 15.04 | 1.05 | 1650.24 | >2000.00 | 216.28 | 209.32 |
| Number | Gene Name | Sequence (5′-3′) |
|---|---|---|
| 1 | hLRP1-3430 | CCUGCAACAAUGGCAGAUGUATT |
| 2 | hLRP1-9620 | GUCCAACUACACGUUACUUAATT |
| 3 | hLRP1-4970 | GCGAACAAACACACUGGCUAATT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, S.; Wang, X.; Han, P.; Lai, Z.; Ren, Y.; Ma, R.; Cheng, C.; Wang, T.; Xu, Y. LRP1-Mediated Endocytosis May Be the Main Reason for the Difference in Cytotoxicity of Curcin and Curcin C on U2OS Osteosarcoma Cells. Toxins 2022, 14, 771. https://doi.org/10.3390/toxins14110771
Qin S, Wang X, Han P, Lai Z, Ren Y, Ma R, Cheng C, Wang T, Xu Y. LRP1-Mediated Endocytosis May Be the Main Reason for the Difference in Cytotoxicity of Curcin and Curcin C on U2OS Osteosarcoma Cells. Toxins. 2022; 14(11):771. https://doi.org/10.3390/toxins14110771
Chicago/Turabian StyleQin, Siying, Xueying Wang, Pan Han, Zhiping Lai, Yingying Ren, Rui Ma, Cheng Cheng, Ting Wang, and Ying Xu. 2022. "LRP1-Mediated Endocytosis May Be the Main Reason for the Difference in Cytotoxicity of Curcin and Curcin C on U2OS Osteosarcoma Cells" Toxins 14, no. 11: 771. https://doi.org/10.3390/toxins14110771





