Type I Toxin-Antitoxin Systems in Clostridia
Abstract
:1. Introduction
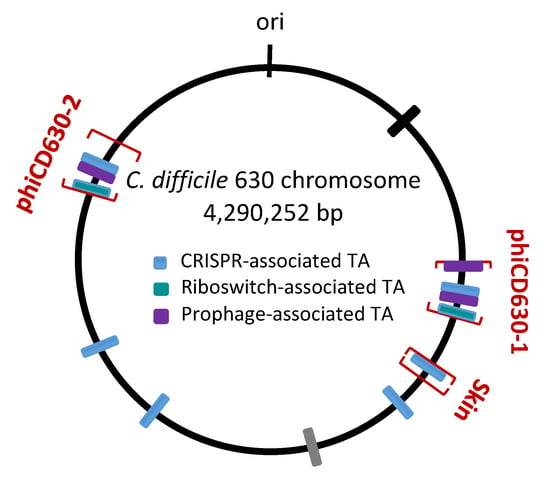
2. Type I TA Systems in Clostridium difficile
3. Comparison with Type I TA Described in Other Gram-Positive Bacteria
4. TA Regulation
5. Potential Functions of Type I TA Systems
6. Possible Applications of Type I TA Systems
7. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Harms, A.; Brodersen, D.E.; Mitarai, N.; Gerdes, K. Toxins, Targets, and Triggers: An Overview of Toxin-Antitoxin Biology. Mol. Cell 2018, 70, 768–784. [Google Scholar] [CrossRef]
- Hayes, F. Toxins-antitoxins: Plasmid maintenance, programmed cell death, and cell cycle arrest. Science 2003, 301, 1496–1499. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Christensen, S.K.; Lobner-Olesen, A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005, 3, 371–382. [Google Scholar] [CrossRef]
- Gerdes, K.; Maisonneuve, E. Bacterial persistence and toxin-antitoxin loci. Annu. Rev. Microbiol. 2012, 66, 103–123. [Google Scholar] [CrossRef] [PubMed]
- Page, R.; Peti, W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016, 12, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Brantl, S.; Jahn, N. sRNAs in bacterial type I and type III toxin-antitoxin systems. FEMS Microbiol. Rev. 2015, 39, 413–427. [Google Scholar] [CrossRef] [Green Version]
- Fineran, P.C.; Blower, T.R.; Foulds, I.J.; Humphreys, D.P.; Lilley, K.S.; Salmond, G.P. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. USA 2009, 106, 894–899. [Google Scholar] [CrossRef]
- Brantl, S. Bacterial type I toxin-antitoxin systems. RNA Biol. 2012, 9, 1488–1490. [Google Scholar] [CrossRef] [Green Version]
- Fozo, E.M.; Makarova, K.S.; Shabalina, S.A.; Yutin, N.; Koonin, E.V.; Storz, G. Abundance of type I toxin-antitoxin systems in bacteria: Searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010, 38, 3743–3759. [Google Scholar] [CrossRef]
- Carroll, K.C.; Bartlett, J.G. Biology of Clostridium difficile: Implications for epidemiology and diagnosis. Annu. Rev. Microbiol. 2011, 65, 501–521. [Google Scholar] [CrossRef]
- Rupnik, M.; Wilcox, M.H.; Gerding, D.N. Clostridium difficile infection: New developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 2009, 7, 526–536. [Google Scholar] [CrossRef] [PubMed]
- Valiente, E.; Cairns, M.D.; Wren, B.W. The Clostridium difficile PCR ribotype 027 lineage: A pathogen on the move. Clin. Microbiol Infect. 2014, 20, 396–404. [Google Scholar] [CrossRef]
- Seekatz, A.M.; Young, V.B. Clostridium difficile and the microbiota. J. Clin. Invest. 2014, 124, 4182–4189. [Google Scholar] [CrossRef] [PubMed]
- Just, I.; Selzer, J.; Wilm, M.; von Eichel-Streiber, C.; Mann, M.; Aktories, K. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature 1995, 375, 500–503. [Google Scholar] [CrossRef]
- Vedantam, G.; Clark, A.; Chu, M.; McQuade, R.; Mallozzi, M.; Viswanathan, V. Clostridium difficile infection: Toxins and non-toxin virulence factors, and their contributions to disease establishment and host response. Gut Microbes 2012, 3, 121–134. [Google Scholar] [CrossRef]
- Janoir, C. Virulence factors of Clostridium difficile and their role during infection. Anaerobe 2016, 37, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Smits, W.K.; Lyras, D.; Lacy, D.B.; Wilcox, M.H.; Kuijper, E.J. Clostridium difficile infection. Nat. Rev. Dis. Primers 2016, 2, 16020. [Google Scholar] [CrossRef] [PubMed]
- Deakin, L.J.; Clare, S.; Fagan, R.P.; Dawson, L.F.; Pickard, D.J.; West, M.R.; Wren, B.W.; Fairweather, N.F.; Dougan, G.; Lawley, T.D. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect. Immun. 2012, 80, 2704–2711. [Google Scholar] [CrossRef] [PubMed]
- Bruggemann, H.; Brzuszkiewicz, E.; Chapeton-Montes, D.; Plourde, L.; Speck, D.; Popoff, M.R. Genomics of Clostridium tetani. Res. Microbiol. 2015, 166, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Blower, T.R.; Short, F.L.; Rao, F.; Mizuguchi, K.; Pei, X.Y.; Fineran, P.C.; Luisi, B.F.; Salmond, G.P. Identification and classification of bacterial Type III toxin-antitoxin systems encoded in chromosomal and plasmid genomes. Nucleic Acids Res. 2012, 40, 6158–6173. [Google Scholar] [CrossRef]
- Rothenbacher, F.P.; Suzuki, M.; Hurley, J.M.; Montville, T.J.; Kirn, T.J.; Ouyang, M.; Woychik, N.A. Clostridium difficile MazF toxin exhibits selective, not global, mRNA cleavage. J. Bacterial. 2012, 194, 3464–3474. [Google Scholar] [CrossRef] [PubMed]
- Gil, F.; Pizarro-Guajardo, M.; Alvarez, R.; Garavaglia, M.; Paredes-Sabja, D. Clostridium difficile recurrent infection: Possible implication of TA systems. Future Microbiol. 2015, 10, 1649–1657. [Google Scholar] [CrossRef]
- Maikova, A.; Peltier, J.; Boudry, P.; Hajnsdorf, E.; Kint, N.; Monot, M.; Poquet, I.; Martin-Verstraete, I.; Dupuy, B.; Soutourina, O. Discovery of new type I toxin-antitoxin systems adjacent to CRISPR arrays in Clostridium difficile. Nucleic Acids Res. 2018, 46, 4733–4751. [Google Scholar] [CrossRef] [PubMed]
- Peltier, J.; Hamiot, A.; Garneau, J.; Boudry, P.; Maikova, A.; Fortier, L.C.; Dupuy, B.; Soutourina, O. Type I toxin-antitoxin systems stabilize prophage regions in the human pathogen Clostridium difficile. in preparation.
- Soutourina, O. RNA-based control mechanisms of Clostridium difficile. Curr. Opin. Microbiol. 2017, 36, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Soutourina, O.A.; Monot, M.; Boudry, P.; Saujet, L.; Pichon, C.; Sismeiro, O.; Semenova, E.; Severinov, K.; Le Bouguenec, C.; Coppee, J.Y.; et al. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet. 2013, 9, e1003493. [Google Scholar] [CrossRef]
- Boudry, P.; Semenova, E.; Monot, M.; Datsenko, K.A.; Lopatina, A.; Sekulovic, O.; Ospina-Bedoya, M.; Fortier, L.C.; Severinov, K.; Dupuy, B.; et al. Function of the CRISPR-Cas System of the Human Pathogen Clostridium difficile. MBio 2015, 6, e01112–e01115. [Google Scholar] [CrossRef] [PubMed]
- Maikova, A.; Severinov, K.; Soutourina, O. New Insights Into Functions and Possible Applications of Clostridium difficile CRISPR-Cas System. Front. Microbiol. 2018, 9, 1740. [Google Scholar] [CrossRef]
- Marraffini, L.A. CRISPR-Cas immunity in prokaryotes. Nature 2015, 526, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Zhang, F. Coupling immunity and programmed cell suicide in prokaryotes: Life-or-death choices. BioEssays 2017, 39, 1–9. [Google Scholar] [CrossRef]
- Silvaggi, J.M.; Perkins, J.B.; Losick, R. Small untranslated RNA antitoxin in Bacillus subtilis. J. Bacteriol. 2005, 187, 6641–6650. [Google Scholar] [CrossRef]
- Fozo, E.M. New type I toxin-antitoxin families from “wild” and laboratory strains of E. coli: Ibs-Sib, ShoB-OhsC and Zor-Orz. RNA Biol. 2012, 9, 1504–1512. [Google Scholar] [CrossRef] [PubMed]
- Fozo, E.M.; Kawano, M.; Fontaine, F.; Kaya, Y.; Mendieta, K.S.; Jones, K.L.; Ocampo, A.; Rudd, K.E.; Storz, G. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 2008, 70, 1076–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iqbal, N.; Guerout, A.M.; Krin, E.; Le Roux, F.; Mazel, D. Comprehensive Functional Analysis of the 18 Vibrio cholerae N16961 Toxin-Antitoxin Systems Substantiates Their Role in Stabilizing the Superintegron. J. Bacteriol. 2015, 197, 2150–2159. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Won, D.; Fozo, E.M. The ZorO-OrzO type I toxin-antitoxin locus: Repression by the OrzO antitoxin. Nucleic Acids Res. 2014, 42, 1930–1946. [Google Scholar] [CrossRef] [PubMed]
- Walling, L.R.; Butler, J.S. Structural Determinants for Antitoxin Identity and Insulation of Cross Talk between Homologous Toxin-Antitoxin Systems. J. Bacteriol. 2016, 198, 3287–3295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, M.; Gao, C.; Wang, Y.; Zhang, H.; He, Z.G. Characterization of the interaction and cross-regulation of three Mycobacterium tuberculosis RelBE modules. PloS ONE 2010, 5, e10672. [Google Scholar] [CrossRef]
- Zhu, L.; Sharp, J.D.; Kobayashi, H.; Woychik, N.A.; Inouye, M. Noncognate Mycobacterium tuberculosis toxin-antitoxins can physically and functionally interact. J. Biol. Chem. 2010, 285, 39732–39738. [Google Scholar] [CrossRef] [PubMed]
- Jahn, N.; Brantl, S.; Strahl, H. Against the mainstream: The membrane-associated type I toxin BsrG from Bacillus subtilis interferes with cell envelope biosynthesis without increasing membrane permeability. Mol. Microbiol. 2015, 98, 651–666. [Google Scholar] [CrossRef]
- Sayed, N.; Nonin-Lecomte, S.; Rety, S.; Felden, B. Functional and structural insights of a Staphylococcus aureus apoptotic-like membrane peptide from a toxin-antitoxin module. J. Biol. Chem. 2012, 287, 43454–43463. [Google Scholar] [CrossRef] [PubMed]
- Grissa, I.; Vergnaud, G.; Pourcel, C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinforma. 2007, 8, 172. [Google Scholar] [CrossRef] [PubMed]
- Durand, S.; Gilet, L.; Condon, C. The essential function of B. subtilis RNase III is to silence foreign toxin genes. PLoS Genet. 2012, 8, e1003181. [Google Scholar] [CrossRef]
- Durand, S.; Jahn, N.; Condon, C.; Brantl, S. Type I toxin-antitoxin systems in Bacillus subtilis. RNA Biol. 2012, 9, 1491–1497. [Google Scholar] [CrossRef]
- Sebaihia, M.; Wren, B.W.; Mullany, P.; Fairweather, N.F.; Minton, N.; Stabler, R.; Thomson, N.R.; Roberts, A.P.; Cerdeno-Tarraga, A.M.; Wang, H.; et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 2006, 38, 779–786. [Google Scholar] [CrossRef]
- Brielle, R.; Pinel-Marie, M.L.; Felden, B. Linking bacterial type I toxins with their actions. Curr. Opin. Microbiol. 2016, 30, 114–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jahn, N.; Brantl, S. One antitoxin--two functions: SR4 controls toxin mRNA decay and translation. Nucleic Acids Res. 2013, 41, 9870–9880. [Google Scholar] [CrossRef] [PubMed]
- Jahn, N.; Brantl, S. Heat-shock-induced refolding entails rapid degradation of bsrG toxin mRNA by RNases Y and J1. Microbiology 2016, 162, 590–599. [Google Scholar] [CrossRef]
- Jahn, N.; Preis, H.; Wiedemann, C.; Brantl, S. BsrG/SR4 from Bacillus subtilis--the first temperature-dependent type I toxin-antitoxin system. Mol. Microbiol. 2012, 83, 579–598. [Google Scholar] [CrossRef]
- Meissner, C.; Jahn, N.; Brantl, S. In Vitro Characterization of the Type I Toxin-Antitoxin System bsrE/SR5 from Bacillus subtilis. J. Biol. Chem. 2016, 291, 560–571. [Google Scholar] [CrossRef]
- Muller, P.; Jahn, N.; Ring, C.; Maiwald, C.; Neubert, R.; Meissner, C.; Brantl, S. A multistress responsive type I toxin-antitoxin system: bsrE/SR5 from the B. subtilis chromosome. RNA Biol. 2016, 13, 511–523. [Google Scholar] [CrossRef]
- Reif, C.; Loser, C.; Brantl, S. Bacillus subtilis Type I antitoxin SR6 Promotes Degradation of Toxin yonT mRNA and Is Required to Prevent Toxic yoyJ Overexpression. Toxins 2018, 10, 74. [Google Scholar] [CrossRef]
- Wang, X.; Wood, T.K. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 2011, 77, 5577–5583. [Google Scholar] [CrossRef]
- Germain-Amiot, N.; Augagneur, Y.; Camberlein, E.; Nicolas, I.; Lecureur, V.; Rouillon, A.; Felden, B. A novel Staphylococcus aureus cis-trans type I toxin-antitoxin module with dual effects on bacteria and host cells. Nucleic Acids Res. 2019, 47, 1759–1773. [Google Scholar] [CrossRef] [PubMed]
- Kawano, M. Divergently overlapping cis-encoded antisense RNA regulating toxin-antitoxin systems from E. coli: Hok/sok, ldr/rdl, symE/symR. RNA Biol. 2012, 9, 1520–1527. [Google Scholar] [CrossRef]
- Kawano, M.; Aravind, L.; Storz, G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 2007, 64, 738–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, J.; Fozo, E.M. sRNA antitoxins: More than one way to repress a toxin. Toxins 2014, 6, 2310–2335. [Google Scholar] [CrossRef] [PubMed]
- Donegan, N.P.; Cheung, A.L. Regulation of the mazEF toxin-antitoxin module in Staphylococcus aureus and its impact on sigB expression. J. Bacteriol. 2009, 191, 2795–2805. [Google Scholar] [CrossRef] [PubMed]
- Pinel-Marie, M.L.; Brielle, R.; Felden, B. Dual toxic-peptide-coding Staphylococcus aureus RNA under antisense regulation targets host cells and bacterial rivals unequally. Cell Rep. 2014, 7, 424–435. [Google Scholar] [CrossRef] [PubMed]
- Riffaud, C.; Pinel-Marie, M.L.; Pascreau, G.; Felden, B. Functionality and cross-regulation of the four SprG/SprF type I toxin-antitoxin systems in Staphylococcus aureus. Nucleic Acids Res. 2019, 47, 1740–1758. [Google Scholar] [CrossRef]
- Sayed, N.; Jousselin, A.; Felden, B. A cis-antisense RNA acts in trans in Staphylococcus aureus to control translation of a human cytolytic peptide. Nat. Struct. Mol. Biol. 2011, 19, 105–112. [Google Scholar] [CrossRef]
- Bronsard, J.; Pascreau, G.; Sassi, M.; Mauro, T.; Augagneur, Y.; Felden, B. sRNA and cis-antisense sRNA identification in Staphylococcus aureus highlights an unusual sRNA gene cluster with one encoding a secreted peptide. Sci. Rep. 2017, 7, 4565. [Google Scholar] [CrossRef]
- Solecki, O.; Mosbah, A.; Baudy Floc’h, M.; Felden, B. Converting a Staphylococcus aureus toxin into effective cyclic pseudopeptide antibiotics. Chem. Biol. 2015, 22, 329–335. [Google Scholar] [CrossRef]
- Wessner, F.; Lacoux, C.; Goeders, N.; Fouquier d’Herouel, A.; Matos, R.; Serror, P.; Van Melderen, L.; Repoila, F. Regulatory crosstalk between type I and type II toxin-antitoxin systems in the human pathogen Enterococcus faecalis. RNA Biol. 2015, 12, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Harp, J.R.; Fozo, E.M. The 5’ UTR of the type I toxin ZorO can both inhibit and enhance translation. Nucleic Acids Res. 2016. [Google Scholar] [CrossRef]
- Arnion, H.; Korkut, D.N.; Masachis Gelo, S.; Chabas, S.; Reignier, J.; Iost, I.; Darfeuille, F. Mechanistic insights into type I toxin antitoxin systems in Helicobacter pylori: The importance of mRNA folding in controlling toxin expression. Nucleic Acids Res. 2017. [Google Scholar] [CrossRef]
- Berghoff, B.A.; Wagner, E.G.H. RNA-based regulation in type I toxin-antitoxin systems and its implication for bacterial persistence. Curr. Genet. 2017, 63, 1011–1016. [Google Scholar] [CrossRef] [PubMed]
- Kristiansen, K.I.; Weel-Sneve, R.; Booth, J.A.; Bjoras, M. Mutually exclusive RNA secondary structures regulate translation initiation of DinQ in Escherichia coli. RNA 2016, 22, 1739–1749. [Google Scholar] [CrossRef]
- Gerdes, K.; Gultyaev, A.P.; Franch, T.; Pedersen, K.; Mikkelsen, N.D. Antisense RNA-regulated programmed cell death. Annu. Rev. Genet. 1997, 31, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.; Luisi, B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011, 9, 578–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Quiroga, C.; Chen, Q.; McAnulty, M.J.; Benedik, M.J.; Wood, T.K.; Wang, X. RalR (a DNase) and RalA (a small RNA) form a type I toxin-antitoxin system in Escherichia coli. Nucleic Acids Res. 2014, 42, 6448–6462. [Google Scholar] [CrossRef] [PubMed]
- Dambach, M.; Irnov, I.; Winkler, W.C. Association of RNAs with Bacillus subtilis Hfq. PloS ONE 2013, 8, e55156. [Google Scholar] [CrossRef]
- Boudry, P.; Gracia, C.; Monot, M.; Caillet, J.; Saujet, L.; Hajnsdorf, E.; Dupuy, B.; Martin-Verstraete, I.; Soutourina, O. Pleiotropic role of the RNA chaperone protein Hfq in the human pathogen Clostridium difficile. J. Bacteriol. 2014, 196, 3234–3248. [Google Scholar] [CrossRef]
- Wozniak, R.A.; Waldor, M.K. A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 2009, 5, e1000439. [Google Scholar] [CrossRef]
- Yao, J.; Guo, Y.; Wang, P.; Zeng, Z.; Li, B.; Tang, K.; Liu, X.; Wang, X. Type II toxin/antitoxin system ParESO/CopASO stabilizes prophage CP4So in Shewanella oneidensis. Environ. Microbiol. 2018, 20, 1224–1239. [Google Scholar] [CrossRef]
- Szekeres, S.; Dauti, M.; Wilde, C.; Mazel, D.; Rowe-Magnus, D.A. Chromosomal toxin-antitoxin loci can diminish large-scale genome reductions in the absence of selection. Mol. Microbiol. 2007, 63, 1588–1605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortier, L.C. Bacteriophages Contribute to Shaping Clostridioides (Clostridium) difficile Species. Front. Microbiol. 2018, 9, 2033. [Google Scholar] [CrossRef]
- Fortier, L.C.; Sekulovic, O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 2013, 4, 354–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Govind, R.; Vediyappan, G.; Rolfe, R.D.; Dupuy, B.; Fralick, J.A. Bacteriophage-mediated toxin gene regulation in Clostridium difficile. J. Virol. 2009, 83, 12037–12045. [Google Scholar] [CrossRef] [PubMed]
- Sekulovic, O.; Meessen-Pinard, M.; Fortier, L.C. Prophage-stimulated toxin production in Clostridium difficile NAP1/027 lysogens. J. Bacteriol. 2011, 193, 2726–2734. [Google Scholar] [CrossRef]
- Dy, R.L.; Przybilski, R.; Semeijn, K.; Salmond, G.P.; Fineran, P.C. A widespread bacteriophage abortive infection system functions through a Type IV toxin-antitoxin mechanism. Nucleic Acids Res. 2014, 42, 4590–4605. [Google Scholar] [CrossRef] [PubMed]
- Alawneh, A.M.; Qi, D.; Yonesaki, T.; Otsuka, Y. An ADP-ribosyltransferase Alt of bacteriophage T4 negatively regulates the Escherichia coli MazF toxin of a toxin-antitoxin module. Mol. Microbiol. 2016, 99, 188–198. [Google Scholar] [CrossRef]
- Koga, M.; Otsuka, Y.; Lemire, S.; Yonesaki, T. Escherichia coli rnlA and rnlB compose a novel toxin-antitoxin system. Genetics 2011, 187, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Pecota, D.C.; Wood, T.K. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J. Bacteriol. 1996, 178, 2044–2050. [Google Scholar] [CrossRef] [PubMed]
- Quax, T.E.; Voet, M.; Sismeiro, O.; Dillies, M.A.; Jagla, B.; Coppee, J.Y.; Sezonov, G.; Forterre, P.; van der Oost, J.; Lavigne, R.; et al. Massive activation of archaeal defense genes during viral infection. J. Virol. 2013, 87, 8419–8428. [Google Scholar] [CrossRef] [PubMed]
- Dorr, T.; Lewis, K.; Vulic, M. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet. 2009, 5, e1000760. [Google Scholar] [CrossRef]
- Dorr, T.; Vulic, M.; Lewis, K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010, 8, e1000317. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.A.; Gollan, B.; Helaine, S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017, 15, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Maisonneuve, E.; Gerdes, K. Molecular mechanisms underlying bacterial persisters. Cell 2014, 157, 539–548. [Google Scholar] [CrossRef]
- Koyanagi, S.; Levesque, C.M. Characterization of a Streptococcus mutans intergenic region containing a small toxic peptide and its cis-encoded antisense small RNA antitoxin. PloS ONE 2013, 8, e54291. [Google Scholar] [CrossRef]
- Maeda, T.; Tanaka, Y.; Inui, M. Glutamine-rich toxic proteins GrtA, GrtB and GrtC together with the antisense RNA AsgR constitute a toxin-antitoxin-like system in Corynebacterium glutamicum. Mol. Microbiol. 2018, 108, 578–594. [Google Scholar] [CrossRef]
- Wen, Y.; Behiels, E.; Devreese, B. Toxin-Antitoxin systems: Their role in persistence, biofilm formation, and pathogenicity. Pathog. Dis. 2014, 70, 240–249. [Google Scholar] [CrossRef]
- Bloom-Ackermann, Z.; Steinberg, N.; Rosenberg, G.; Oppenheimer-Shaanan, Y.; Pollack, D.; Ely, S.; Storzi, N.; Levy, A.; Kolodkin-Gal, I. Toxin-Antitoxin systems eliminate defective cells and preserve symmetry in Bacillus subtilis biofilms. Environ. Microbiol. 2016, 18, 5032–5047. [Google Scholar] [CrossRef] [PubMed]
- Georgiades, K.; Raoult, D. Genomes of the most dangerous epidemic bacteria have a virulence repertoire characterized by fewer genes but more toxin-antitoxin modules. PloS ONE 2011, 6, e17962. [Google Scholar] [CrossRef] [PubMed]
- Lobato-Marquez, D.; Moreno-Cordoba, I.; Figueroa, V.; Diaz-Orejas, R.; Garcia-del Portillo, F. Distinct type I and type II toxin-antitoxin modules control Salmonella lifestyle inside eukaryotic cells. Sci. Rep. 2015, 5, 9374. [Google Scholar] [CrossRef]
- Chan, W.T.; Balsa, D.; Espinosa, M. One cannot rule them all: Are bacterial toxins-antitoxins druggable? FEMS Microbiol. Rev. 2015, 39, 522–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghafourian, S.; Raftari, M.; Sadeghifard, N.; Sekawi, Z. Toxin-antitoxin Systems: Classification, Biological Function and Application in Biotechnology. Curr. Issues. Mol. Biol. 2014, 16, 9–14. [Google Scholar]
- Kang, S.M.; Kim, D.H.; Jin, C.; Lee, B.J. A Systematic Overview of Type II and III Toxin-Antitoxin Systems with a Focus on Druggability. Toxins 2018, 10, 515. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Lee, B.J. Structure, Biology, and Therapeutic Application of Toxin-Antitoxin Systems in Pathogenic Bacteria. Toxins 2016, 8, 305. [Google Scholar] [CrossRef]
- Zhang, X.Z.; Yan, X.; Cui, Z.L.; Hong, Q.; Li, S.P. mazF, a novel counter-selectable marker for unmarked chromosomal manipulation in Bacillus subtilis. Nucleic Acids Res. 2006, 34, e71. [Google Scholar] [CrossRef]
- Wu, J.; Deng, A.; Sun, Q.; Bai, H.; Sun, Z.; Shang, X.; Zhang, Y.; Liu, Q.; Liang, Y.; Liu, S.; et al. Bacterial Genome Editing via a Designed Toxin-Antitoxin Cassette. ACS Synth. Biol. 2018, 7, 822–831. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wang, Y.; Wei, C.; Liu, Q.; Jin, X.; Du, G.; Chen, J.; Kang, Z. A new sRNA-mediated posttranscriptional regulation system for Bacillus subtilis. Biotechnol. Bioeng. 2018, 115, 2986–2995. [Google Scholar] [CrossRef]
- Booth, J.A.; Suganthan, R.; Gaustad, P.; Bjoras, M. Development of DinQ from Escherichia coli as an anti-cell-envelope antibiotic. Int. J. Antimicrob. Agents 2015, 45, 196–197. [Google Scholar] [CrossRef] [PubMed]



| Number | Toxin, Length 1 | Antitoxin RNA | Location | Association | Comment 2 | Ref. |
|---|---|---|---|---|---|---|
| 1 * | CD0440.1, 46AA | CD630_n00150 | ||||
| 2 *,3 | CD0904.1 (CD630_n00350), 35AA | AS CD0904.1 | phiCD630-1 | Prophage stabilization | [24] | |
| 3 *,3 | CD0956.2, 53AA 5 | RCd10 | phiCD630-1 | CRISPR 3/4 | Prophage stabilization | [23,24] |
| 4 | CD0956.3, 34AA 6 | AS CD0956.3 | phiCD630-1 | Prophage stabilization | [24] | |
| 5 *,3 | CD0977.1, 47AA 7 | RCd11 | phiCD630-1 | cdi1_5 | Prophage stabilization | [24] |
| 6 * | CD1233.1, 50AA | SQ808 | skin | CRISPR 6 | [23] | |
| 7 * | CD1418.2, 50AA | CD630_n00500 | CRISPR 7 | [23] | ||
| 8 4 | CD1663.2, 59AA | CD630_n00610 | CRISPR 9 | [23] | ||
| 9 * | CD2299.1, 50AA | SQ1641 | CRISPR 11 | [23] | ||
| 10 *,3 | CD2517.1, 52AA | RCd8 | CRISPR 12 | [23] | ||
| 11 *,3 | CD2889, 47AA 7 | RCd12 | phiCD630-2 | cdi1_4 | Prophage stabilization | [24] |
| 12 *,3 | CD2907.1, 53AA 5 | RCd9 | phiCD630-2 | CRISPR 16/15 | Prophage stabilization | [23,24] |
| 13 | CD2907.2, 34AA 6 | AS CD2907.2 | phiCD630-2 | Prophage stabilization | [24] |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soutourina, O. Type I Toxin-Antitoxin Systems in Clostridia. Toxins 2019, 11, 253. https://doi.org/10.3390/toxins11050253
Soutourina O. Type I Toxin-Antitoxin Systems in Clostridia. Toxins. 2019; 11(5):253. https://doi.org/10.3390/toxins11050253
Chicago/Turabian StyleSoutourina, Olga. 2019. "Type I Toxin-Antitoxin Systems in Clostridia" Toxins 11, no. 5: 253. https://doi.org/10.3390/toxins11050253