Characterization of Human Type C Enterotoxin Produced by Clinical S. epidermidis Isolates
Abstract
:1. Introduction
2. Results
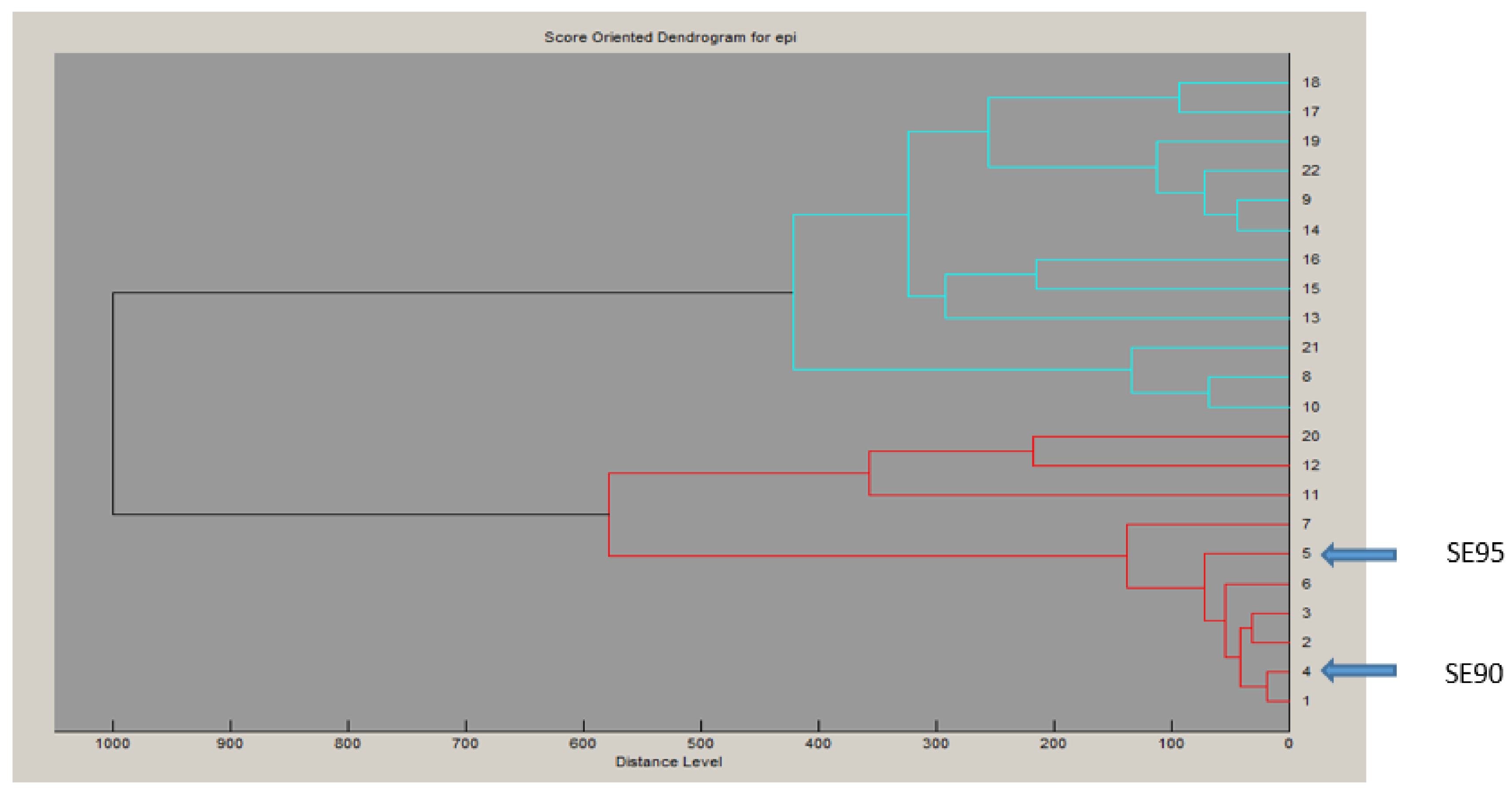
2.1. Characteristics of the Putative secepi Gene Encoded by Clinical S. epidermidis Strains
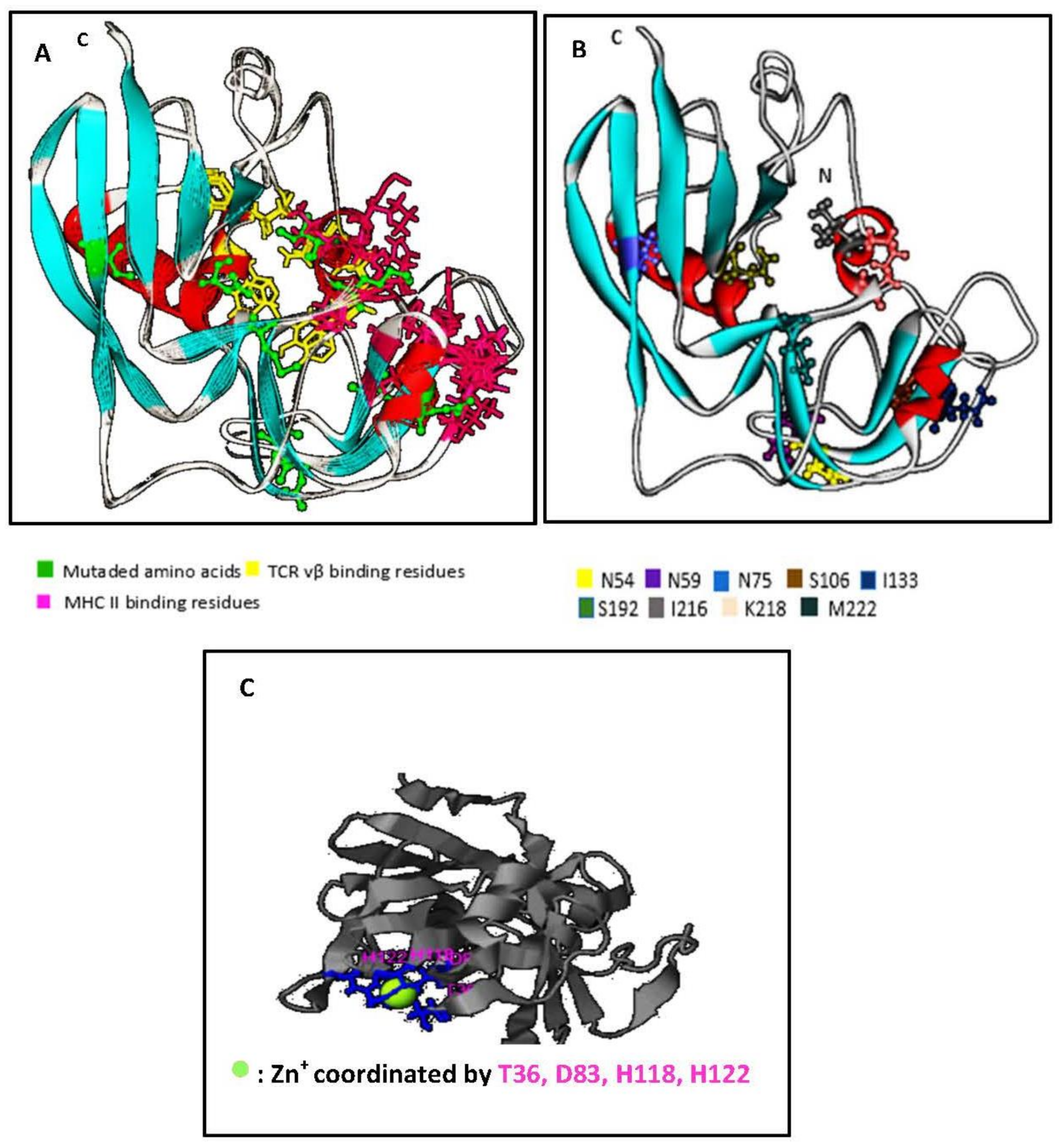
2.2. Structural Features of SECepi and Structural Homology with SECaureus
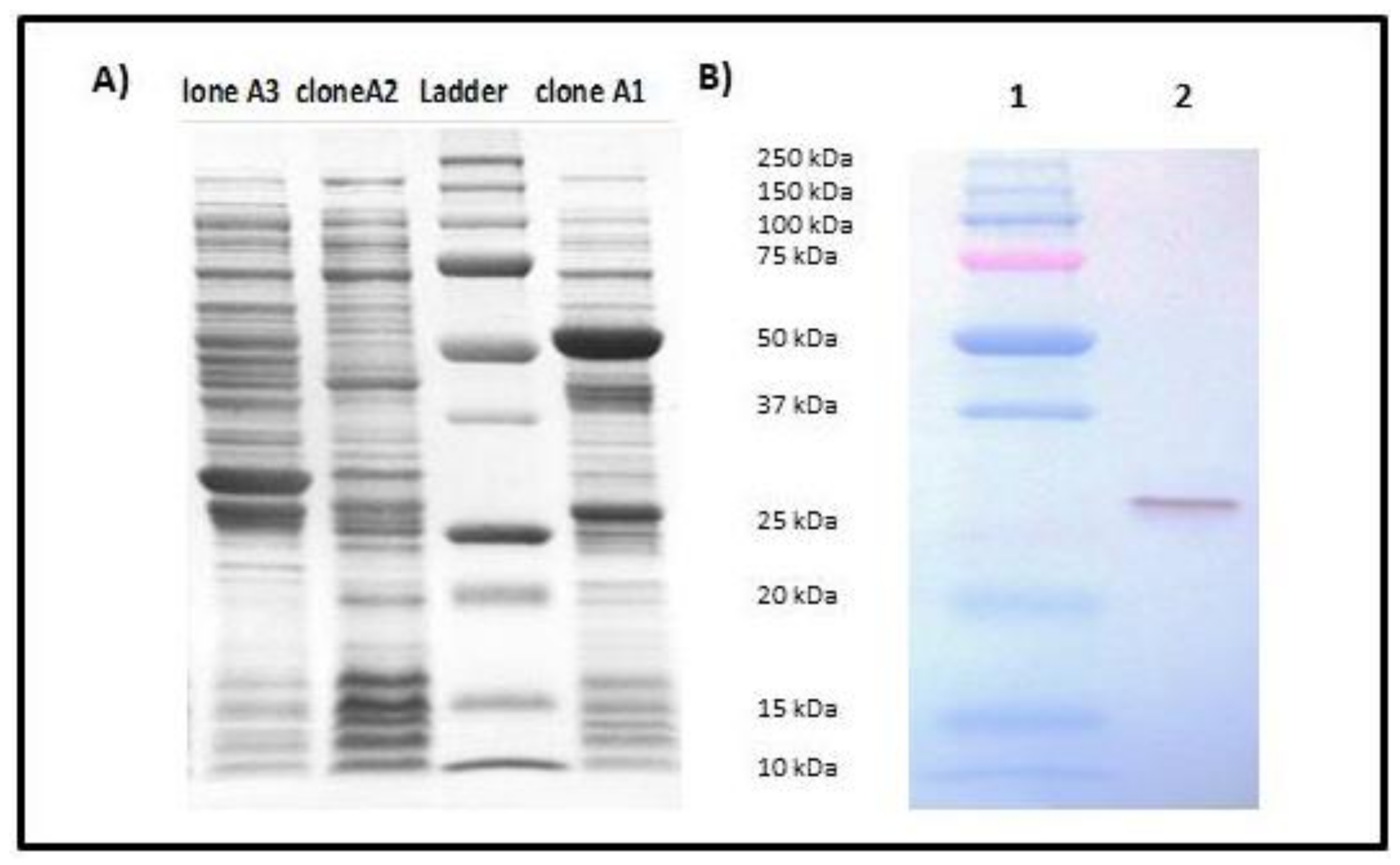
2.3. Expression and Purification of rSECepi
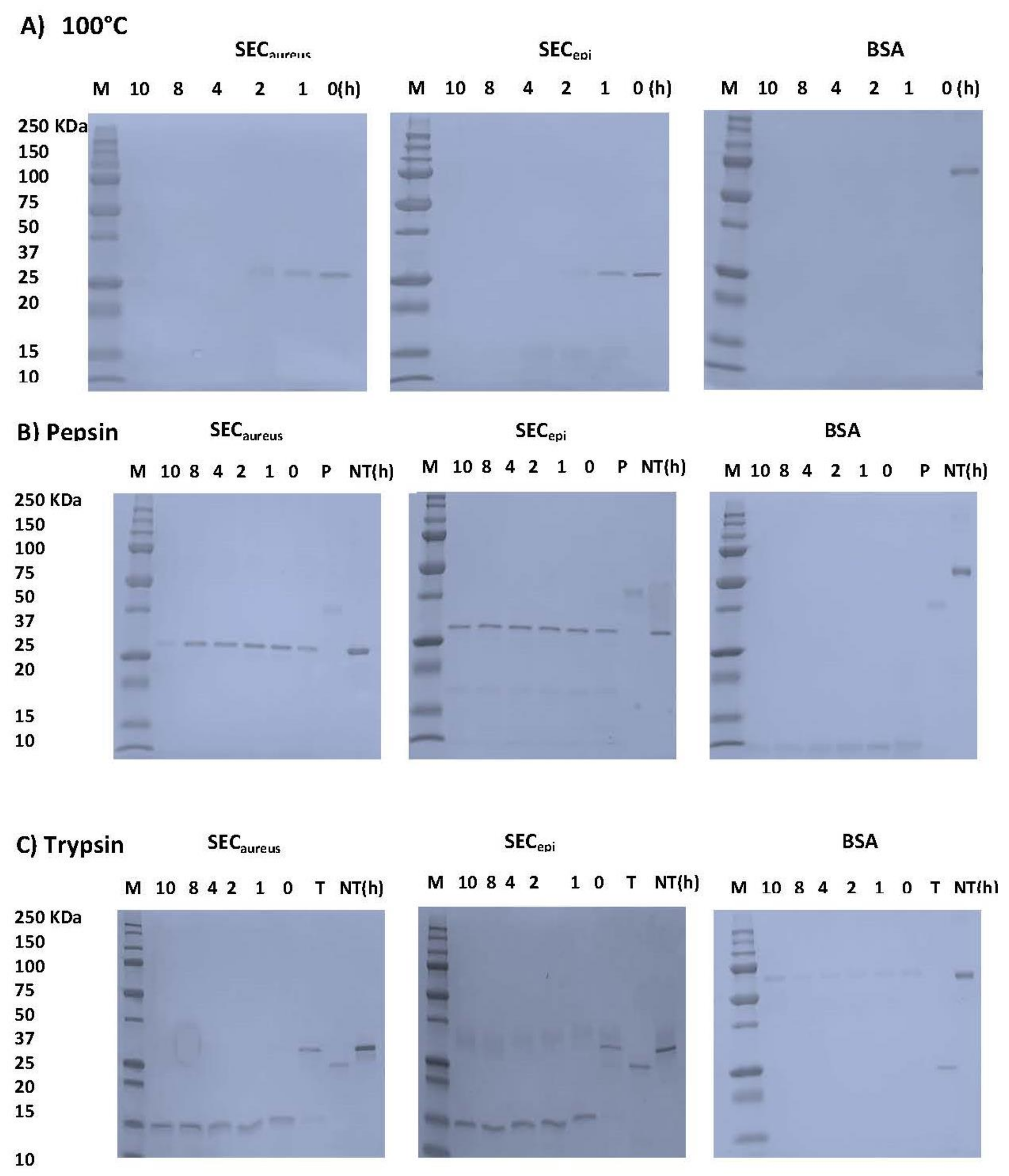
2.4. SECepi Is Stable to Heat Treatment and Digestive Enzymes Treatment
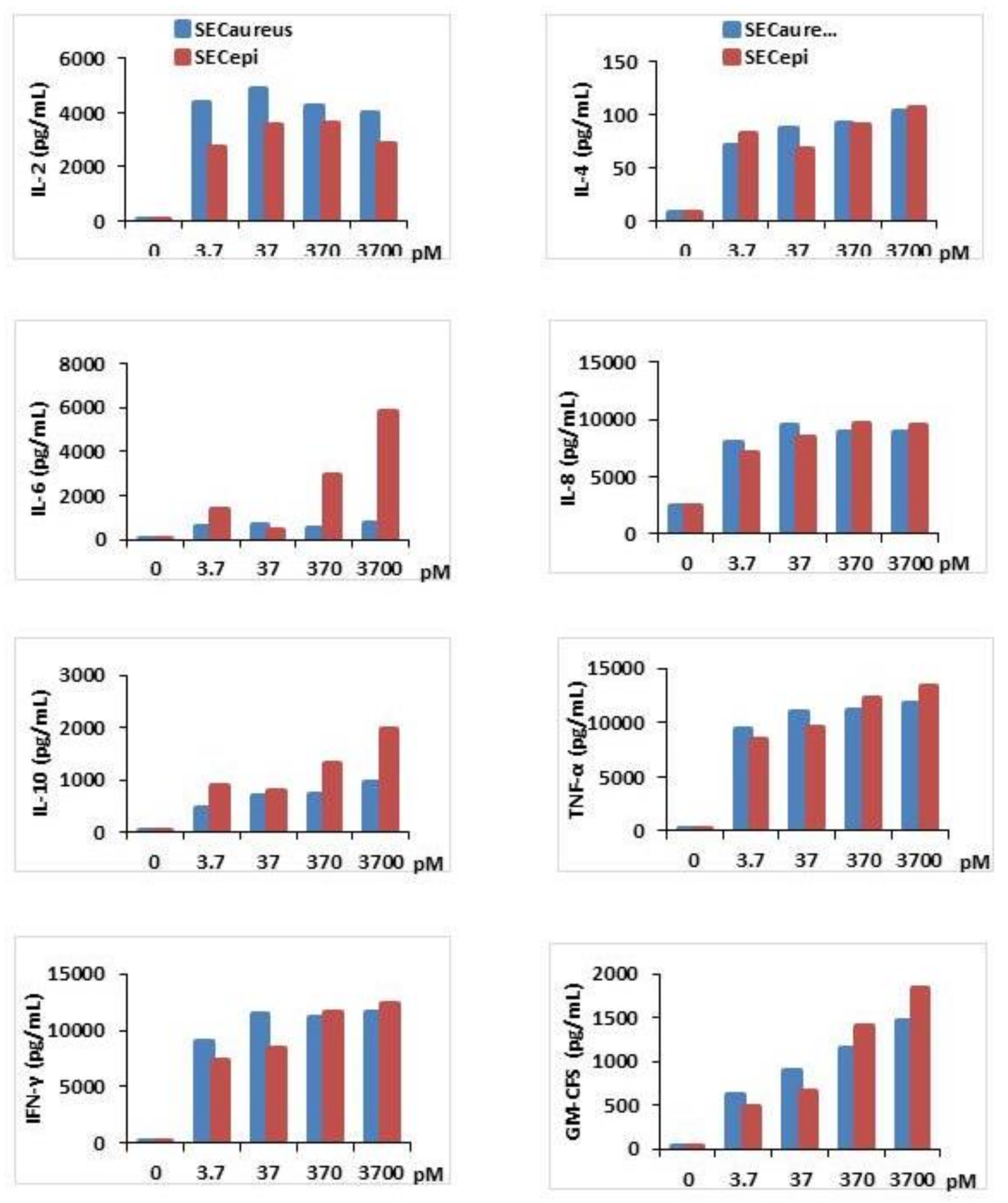
2.5. Functional Analyses of SECepi
3. Discussion
4. Materials and Methods
4.1. Ethical Statement
4.2. Bacterial Strains, Vectors Media, and Growth Conditions
4.3. DNA Isolation
4.4. Nucleotide Sequencing and Analysis
4.5. Bioinformatic Analysis
4.6. Cloning of secepi Gene
4.7. Expression and Purification of rSECepi
4.8. Western Blotting
4.9. Determination of Enzyme and Heat Stability of SECepi
4.10. Human Peripheral Blood Mononuclear Cells (hPMBC) Purification and Culture
4.11. Cell Proliferation Assays
4.11.1. Cell Proliferation with Different Concentrations of SECepi and SECaureus
4.11.2. Cell Proliferation after Toxins SECepi and SECaureus Heating
4.12. Screening for Cytokines Production by Activated T Cells
4.13. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Spaulding, A.; Salgado-Pabón, W.; Kohler, P.; Horswill, A.; Leung, D.; Schlievert, P. Staphylococcal and streptococcal superantigen exotoxins. Clin. Microbiol. Rev. 2013, 26, 422–447. [Google Scholar] [CrossRef] [PubMed]
- Argudín, M.A.; Mendoza, M.C.; Rodicio, M.R. Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2010, 2, 1751–1773. [Google Scholar] [CrossRef] [PubMed]
- Vu, B.G.; Stach, C.S.; Salgado-Pabon, W.; Diekema, D.J.; Gardner, S.E.; Schlievert, P.M. Superantigens of Staphylococcus aureus from patients with diabetic foot ulcers. J. Infect. Dis. 2014, 210, 1920–1927. [Google Scholar] [CrossRef] [PubMed]
- Hennekinne, J.A.; Buyser, M.L.; Dragacci, S. Staphylococcus aureus and its food poisoning toxins: Characterization and outbreak investigation. FEMS Microbiol. Rev. 2012, 36, 815–836. [Google Scholar] [CrossRef] [PubMed]
- Mempel, M.; Lina, G.; Hojka, M.; Schnopp, C.; Seidl, H.P.; Schafer, T.; Ring, J.; Vandenesch, F.; Abeck, D. High prevalence of superantigens associated with the egc locus. In Staphylococcus aureus isolates from patients with atopic eczema. Eur. J. Clin. Microbiol. Infect. Dis. 2003, 22, 306–309. [Google Scholar] [PubMed]
- Ye, Y.M.; Hur, G.Y.; Park, H.J.; Kim, S.H.; Kim, H.M.; Park, H.S. Association of specific IgE to staphylococcal superantigens with the phenotype of chronic urticaria. J. Korean Med. Sci. 2008, 23, 845–851. [Google Scholar] [CrossRef] [PubMed]
- Bunning, V.K.; Lindsay, J.A.; Archer, D.L. Chronic health effects of microbial foodborne disease. World Health Stat. Q. 1997, 50, 51–56. [Google Scholar] [PubMed]
- Salgado-Pabón, W.; Breshears, L.; Spaulding, A.R.; Merriman, J.A.; Stach, C.S.; Horswill, A.R.; Peterson, M.L.; Schlievert, P.M. Superantigens are critical for Staphylococcus aureus infective endocarditis, sepsis, and acute kidney injury. MBio 2013, 4, e00494-13. [Google Scholar] [CrossRef] [PubMed]
- Wilson, G.J.; Seo, K.S.; Cartwright, R.A.; Connelley, T.; Chuang-Smith, O.N.; Merriman, J.A.; Guinane, C.M.; Park, J.Y.; Bohach, G.A.; Schlievert, P.M.; et al. A novel core genome-encoded superantigen contributes to lethality of community-associated MRSA necrotizing pneumonia. PLoS Pathog. 2011, 7, e1002271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinges, M.M.; Orwin, P.M.; Schlievert, P.M. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 2000, 13, 16–34. [Google Scholar] [CrossRef] [PubMed]
- Podkowik, M.; Seo, K.S.; Schubert, J.; Tolo, I.; Robinson, D.A.; Bania, J.; Bystroń, J. Genotype and enterotoxigenicity of Staphylococcus epidermidis isolate from ready to eat meat products. Int. J. Food Microbiol. 2016, 229, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Veras, J.F.; Carmo, L.S.; Tong, L.C.; Shupp, J.W.; Cummings, C.; Santos, D.A.; Cerqueira, M.M.O.; Cantini, A.; Nicoli, J.R.; Jett, M. A study of the enterotoxigenicity of coagulase negative and coagulase-positive staphylococcal isolates from food poisoning outbreaks in Minas Gerais, Brazil. Int. J. Infect. Dis. 2008, 12, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Stach, C.S.; Vu, B.G.; Schlievert, P.M. Determining the presence of superantigens in coagulase negative staphylococci from humans. PLoS ONE 2015, 10, e0143341. [Google Scholar] [CrossRef] [PubMed]
- Sato’o, Y.; Omoe, K.; Ono, H.K.; Nakane, A.; Hu, D.L. A novel comprehensive analysis method for Staphylococcus aureus pathogenicity islands. Microbiol. Immunol. 2013, 57, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Ono, H.K.; Sato’o, Y.; Narita, K.; Naito, I.; Hirose, S.; Hisatsune, J.; Asano, K.; Hu, D.L.; Omoe, K.; Sugai, M.; et al. Identification and characterization of a novel staphylococcal emetic toxin. Appl. Environ. Microbiol. 2015, 81, 7034–7040. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.; Rasooly, A. Staphylococcal enterotoxins. Int. J. Food Microbiol. 2000, 61, 1–10. [Google Scholar] [CrossRef]
- Wieneke, A. Enterotoxin production by strains of Staphylococcus aureus isolated from foods and human beings. J. Hyg. 1974, 73, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Marr, J.C.; Lyon, J.D.; Roberson, J.R.; Lupher, M.; Davis, W.C.; Bohach, G.A. Characterization of novel type C staphylococcal enterotoxins: Biological and evolutionary implications. Infect. Immun. 1993, 61, 4254–4262. [Google Scholar] [PubMed]
- Reiser, R.F.; Robbins, R.N.; Noleto, A.L.; Khoe, G.P.; Bergdoll, M.S. Identification, purification, and some physicochemical properties of staphylococcal enterotoxin C3. Infect. Immun. 1984, 45, 625–630. [Google Scholar] [PubMed]
- Deringer, J.R.; Ely, R.J.; Stauffacher, C.V.; Bohach, G.A. Subtype-specific interactions of type C staphylococcal enterotoxins with the T-cell receptor. Mol. Microbiol. 1996, 22, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Staphylococcus epidermidis—The “accidental” pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Ziebuhr, W. Staphylococcus aureus and Staphylococcus epidermidis: Emerging pathogens in nosocomial infections. Contrib. Microbiol. 2001, 8, 102–107. [Google Scholar] [PubMed]
- Madhusoodanan, J.; Seo, K.S.; Park, J.Y.; Gill, A.L.; Waterhouse, J.; Remortel, B.; Bohach, G.; Gill, S.R. Abstract of 107th General Meeting of the American Society for Microbiology, Microbiol, Toronto, Japan, 21–25, May, 2007; American Society for Microbiology: Washington, DC, USA, 2007; p. B-420. [Google Scholar]
- Nanoukon, C.; Argemi, X.; Sogbo, F.; Orekan, J.; Keller, D.; Affolabi, D.; Schramm, F.; Riegel, P.; Baba-Moussa, L.; Prévost, G. Pathogenic features of clinically significant coagulase-negative staphylococci in hospital and community infections in Benin. Int. J. Med. Microbiol. 2017, 307, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Argemi, X.; Nanoukon, C.; Affolabi, D.; Keller, D.; Hansmann, Y.; Riegel, P.; Baba-Moussa, L.; Prévost, G. Comparative genomics and identification of an enterotoxin-bearing pathogenicity island SEPI-1/SECI-1, in Staphylococcus epidermidis Pathogenic Strains. Toxins (Basel) 2018, 10, E93. [Google Scholar] [CrossRef] [PubMed]
- Madhusoodanan, J.; Seo, K.S.; Remortel, B.; Park, J.Y.; Hwang, S.Y.; Fox, L.K.; Park, Y.H.; Deobald, C.F.; Wang, D.; Liu, S.; et al. An Enterotoxin-bearing pathogenicity island in Staphylococcus epidermidis. J. Bacteriol. 2011, 193, 1854–1862. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.I.; Sadler, I.; Jablonski, L.M.; Callantine, S.D.; Deobald, C.F.; Stauffacher, C.V.; Bohach, G.A. Zinc-mediated dimerization and Its effect on activity and conformation of staphylococcal enterotoxin type C. J. Biol. Chem. 2002, 277, 22839–22846. [Google Scholar] [CrossRef] [PubMed]
- Schlievert, P.M.; Bohach, G.A.; Ohlendorf, D.H.; Stauffacher, C.V.; Leung, D.Y.; Murray, D.L.; Prasad, G.S.; Earhart, C.A.; Jablonski, L.M.; Chi, Y.I. Molecular structure of staphylococcus and streptococcus suerantigens. J. Clin. Immunol. 1995, 15 (Suppl. 6), 4S–10S. [Google Scholar] [CrossRef] [PubMed]
- Attien, P.; Sina, H.; Moussaoui, W.; Zimmermann-Meise, G.; Dadié, T.; Keller, D.; Riegel, P.; Edoh, V.; Kotchoni, S.O.; Diè, M.; et al. Mass spectrometry and multiplex antigen assays to assess microbial quality and toxin production of Staphylococcus aureus strains isolated from clinical and food samples. Biomed. Res. Int. 2014, 485620. [Google Scholar] [CrossRef]
- Li, S.J.; Hu, D.L.; Maina, E.K.; Shinagawa, K.; Omoe, K.; Nakane, A. Superantigenic activity of toxic shock syndrome toxin-1 is resistant to heating and digestive enzymes. J. Appl. Microbiol. 2011, 110, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Mera, S.; Tatulescu, D.; Cismaru, C.; Bondor, C.; Slavcovici, A.; Zanc, V.; Carstina, D.; Oltean, M. Multiplex cytokine profiling in patients with sepsis. Apmis 2011, 119, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chaudhry, H.; Zhong, Y.; Ali, M.; Perkins, L.A.; Owens, W.; Morales, J.E.; McGuire, F.; Zumbrun, E.; Zhang, J.; et al. Dysregulation in microRNA expression in peripheral blood mononuclear cells of sepsis patients is associated with immunopathology. Cytokine 2015, 71, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. Proinflammatory and anti-inflammatory cytokines as mediators in the pathogenesis of septic shock. Chest 1997, 112, 321S–329S. [Google Scholar] [CrossRef] [PubMed]
- Krakauer, T.; Pradhan, K.; Stiles, B.G. Staphylococcal superantigens spark host-mediated danger signals. Front. Immunol. 2016, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lamphear, J.G.; Bohach, G.A.; Rich, R.R. Structural dichotomy of staphylococcal enterotoxin C superantigens leading to MHC class II independent activation of T lymphocytes. J. Immunol. 1998, 160, 2107–2114. [Google Scholar] [PubMed]
- Leder, L.; Llera, A.; Lavoie, P.M.; Lebedeva, M.I.; Li, H.; Sekaly, R.P.; Bohach, G.A.; Gahr, P.J.; Schlievert, P.M.; Karjalainen, K.; et al. A mutational analysis of the binding of staphylococcal enterotoxins B and C3 to the T cell receptor- chain and major histocompatibility complex class II. J. Exp. Med. 1998, 187, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Rossle, S.C.; Bisch, P.M.; Lone, Y.C.; Abastado, J.P.; Kourilsky, P.; Bellio, M. Mutational analysis and molecular modeling of the binding of Staphylococcus aureus enterotoxin C2 to a murine T cell receptor V-10 chain. Eur. J. Immunol. 2002, 32, 2172–2178. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Xu, M.; Cai, Y.; Liu, C.; Su, Z.; Zhang, C. Biological characterization of the zinc site coordinating histidine residues of staphylococcal enterotoxin C2. Microbiology 2009, 155, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Bergdoll, M. Staphylococcus aureus. Foodborne Pathog. Dis. 1991, D1989, 463–524. [Google Scholar]
- Orwin, P.M.; Leung, D.Y.; Tripp, T.J.; Bohach, G.A.; Earhart, C.A.; Ohlendorf, D.H.; Schlievert, P.M. Characterization of a novel staphylococcal enterotoxin-like superantigen, a member of the group V subfamily of pyrogenic toxins. Biochemistry 2002, 41, 14033–14040. [Google Scholar] [CrossRef] [PubMed]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed]
- Translate. Available online: http://web.expasy.org/translate (accessed on 18 February 2017).
- ProtParam. Available online: http://web.expasy.org/protparam (accessed on 21 January 2017).
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Podkowik, M.; Bystroń, J.; Bania, J. Genotypes, antibiotic resistance and virulence factors of staphylococci from ready-to-eat food. Foodborne Pathog. Dis. 2012, 9, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.; Girardot, R.; Piémont, Y.; Prévost, G.; Colin, D.A. Analysis of the specificity of Panton-Valentine leucocidin and gamma-hemolysin F component binding. Infect. Immun. 2009, 7, 266–273. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nanoukon, C.; Affolabi, D.; Keller, D.; Tollo, R.; Riegel, P.; Baba-Moussa, L.; Prévost, G. Characterization of Human Type C Enterotoxin Produced by Clinical S. epidermidis Isolates. Toxins 2018, 10, 139. https://doi.org/10.3390/toxins10040139
Nanoukon C, Affolabi D, Keller D, Tollo R, Riegel P, Baba-Moussa L, Prévost G. Characterization of Human Type C Enterotoxin Produced by Clinical S. epidermidis Isolates. Toxins. 2018; 10(4):139. https://doi.org/10.3390/toxins10040139
Chicago/Turabian StyleNanoukon, Chimène, Dissou Affolabi, Daniel Keller, Rodrigue Tollo, Philippe Riegel, Lamine Baba-Moussa, and Gilles Prévost. 2018. "Characterization of Human Type C Enterotoxin Produced by Clinical S. epidermidis Isolates" Toxins 10, no. 4: 139. https://doi.org/10.3390/toxins10040139




