Assessment of the Nutraceutical Effects of Oleuropein and the Cytotoxic Effects of Adriamycin, When Administered Alone and in Combination, in MG-63 Human Osteosarcoma Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Oleuropein Isolation
2.2. Cell Line
2.3. MTT Cytotoxicity-Proliferation Assay
2.4. Flow Cytometry
2.5. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.6. Western Blotting
2.7. ELISA Method
2.8. Metabolites Extraction and Preparation of NMR (Nuclear Magnetic Resonance) Samples
2.9. NMR Profiling
2.10. Statistical Analysis
3. Results
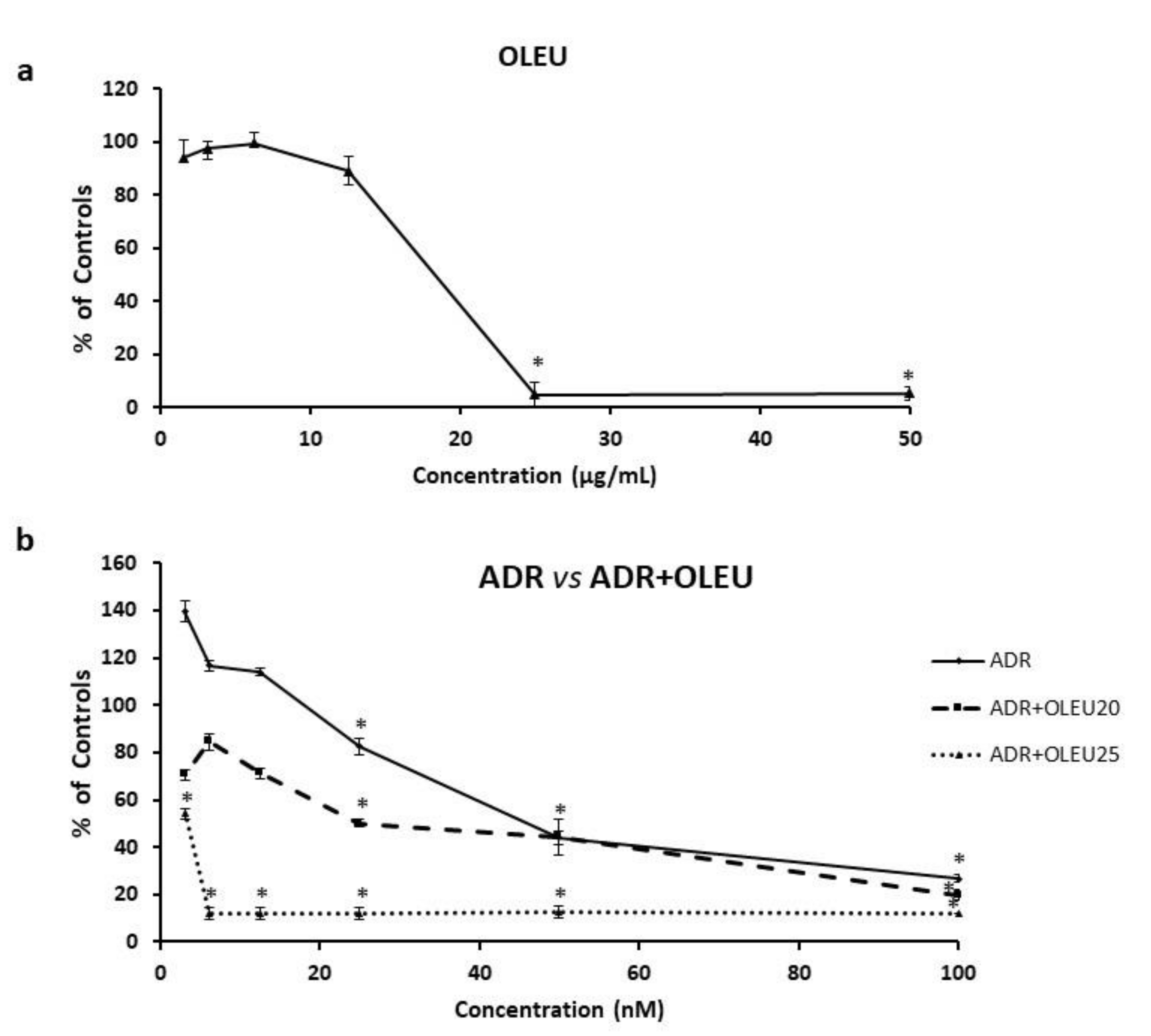
3.1. Cytotoxicity of ADR or/and OLEU
3.2. Alterations in Cell Cycle Distribution
3.3. Autophagy Related Genes’ Expression
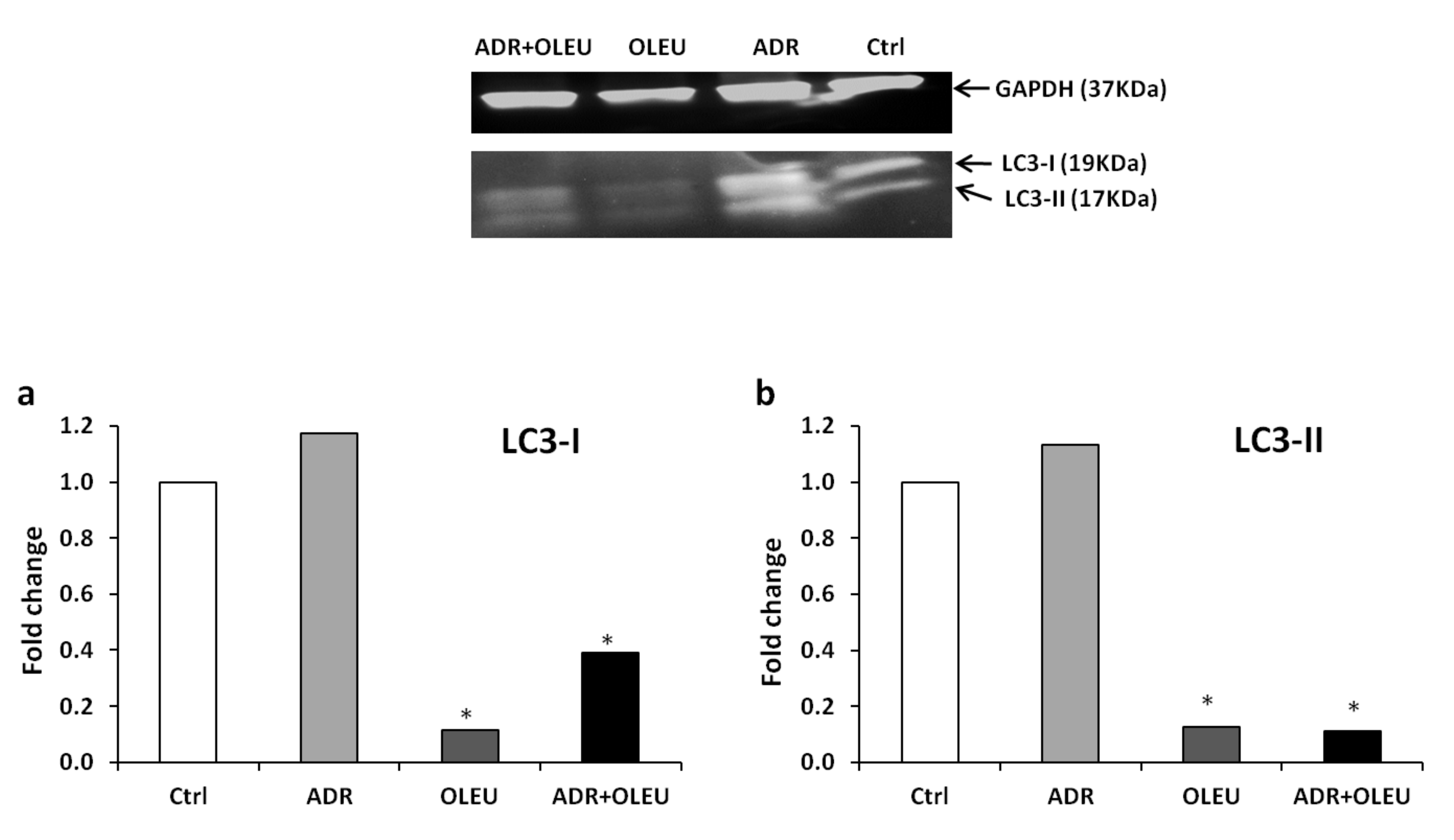
3.4. Expression of LC3 (Microtubule-Associated Protein 1A/1B-Light Chain 3) Proteins
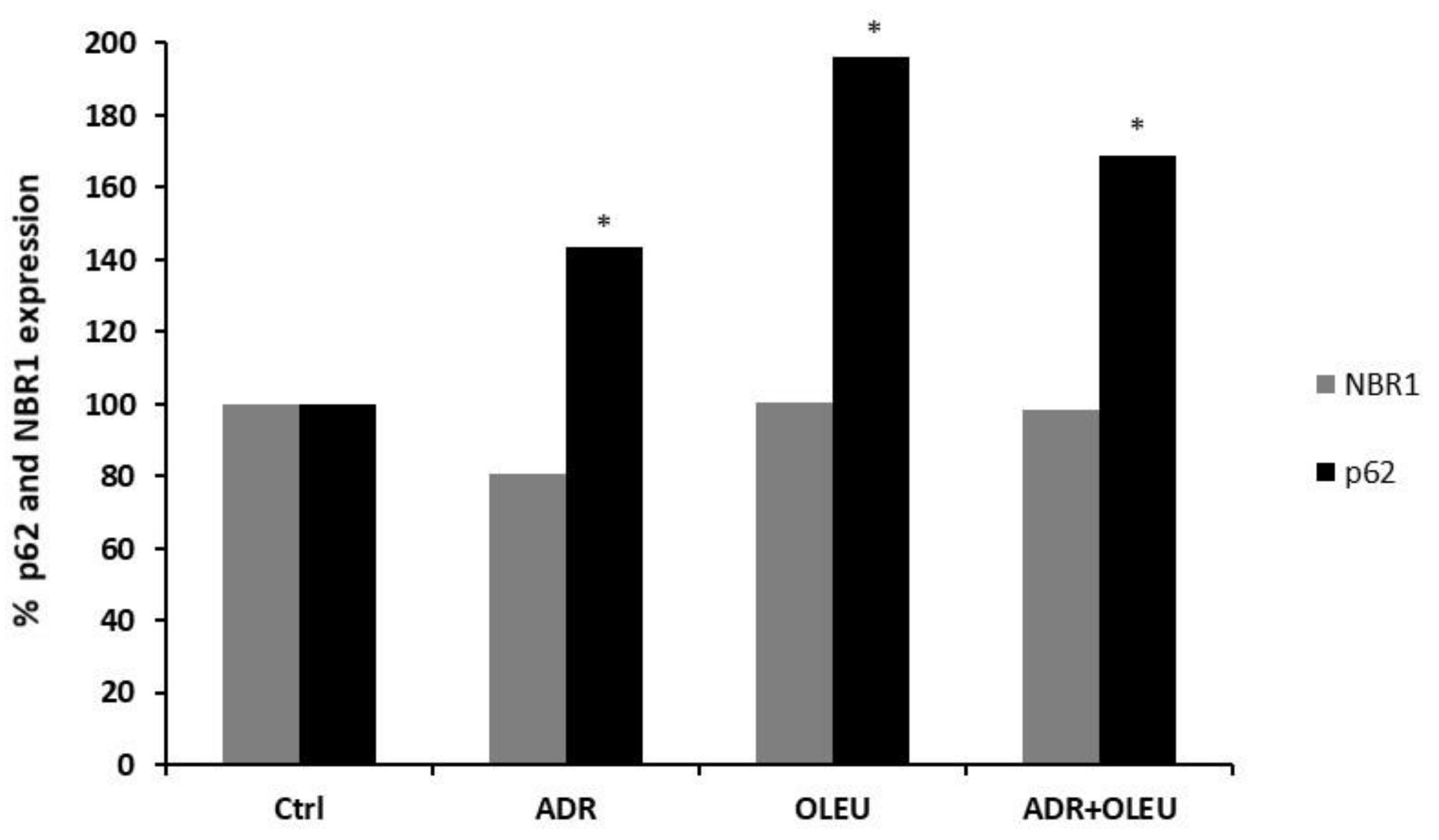
3.5. NBR1 (Neighbour of BRCA1 Gene 1 Protein) and p62 (Ubiquitin-Binding Protein p62) Expression
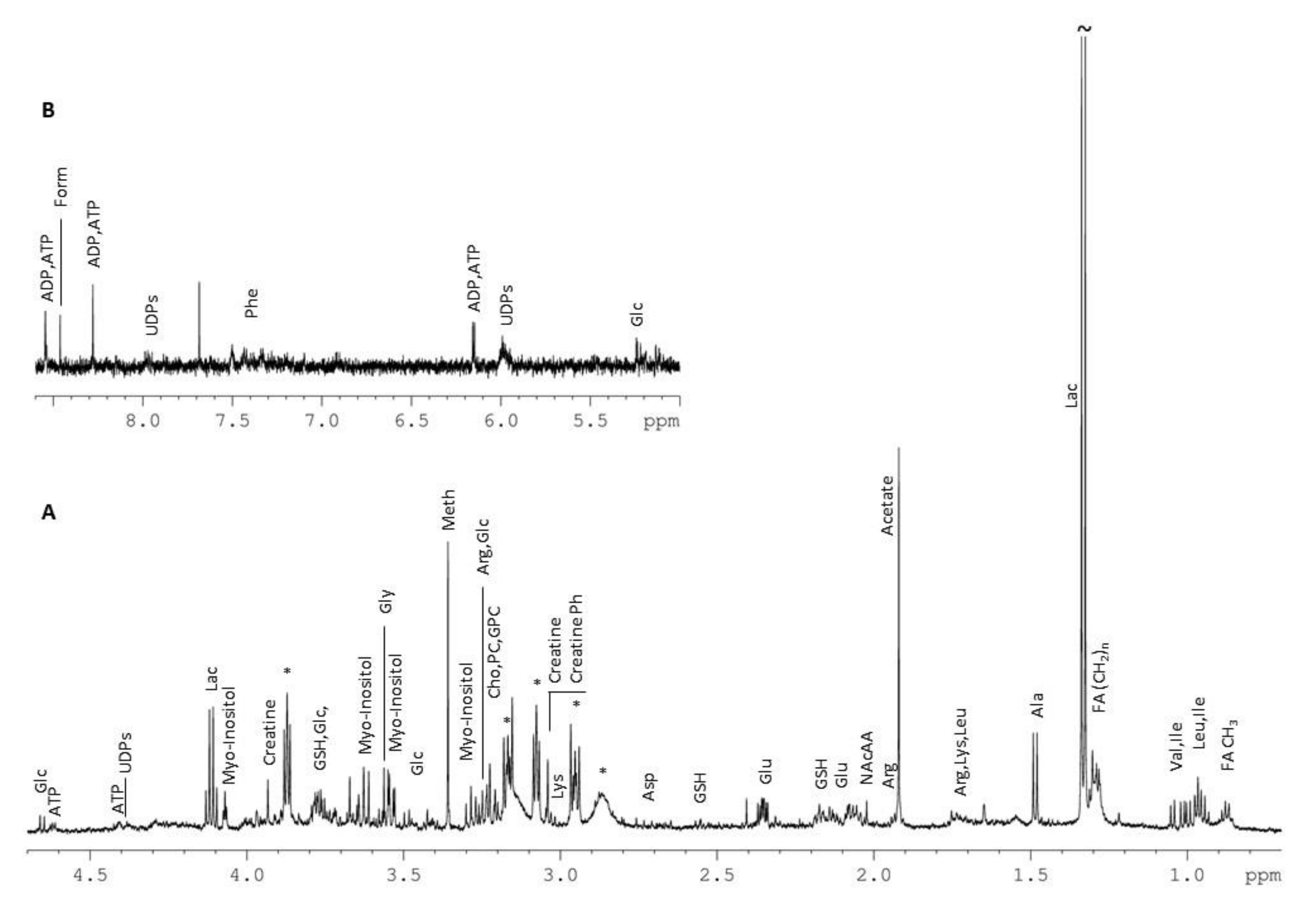
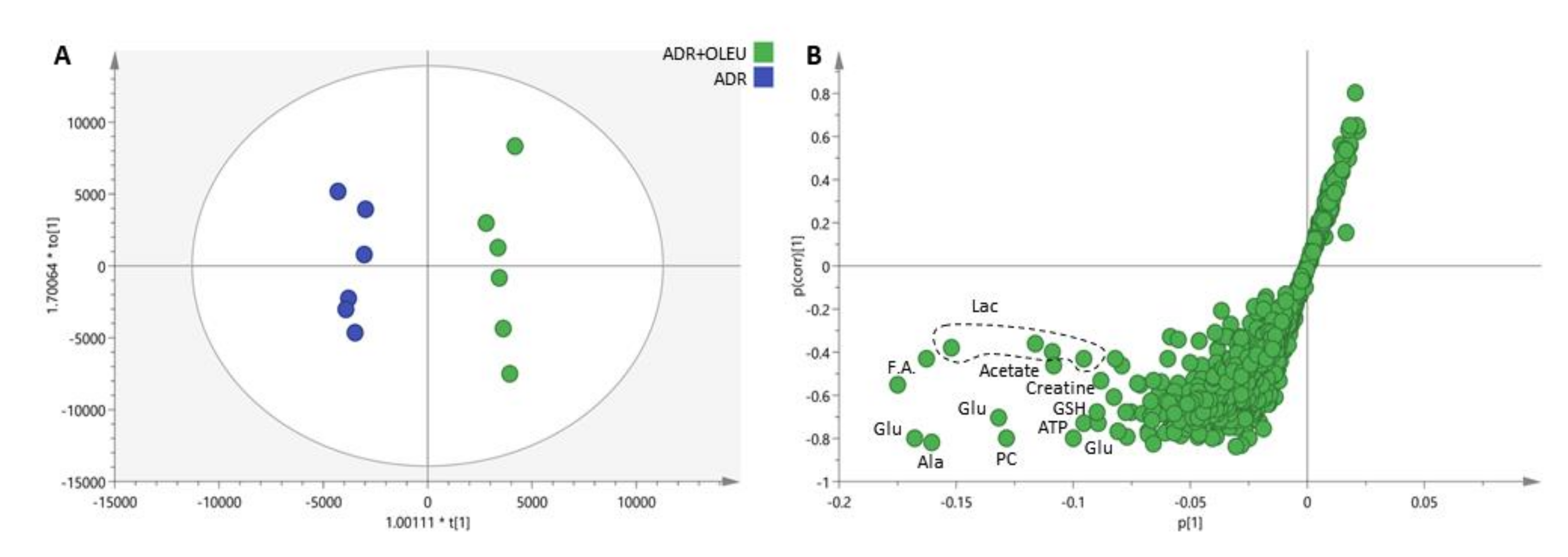
3.6. Metabolic Profiling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bacci, G.; Ferrari, S.; Delepine, N.; Bertoni, F.; Picci, P.; Mercuri, M.; Bacchini, P.; Del Prever, A.B.; Tienghi, A.; Comandone, A.; et al. Predictive factors of histologic response to primary chemotherapy in osteosarcoma of the extremity: Study of 272 patients preoperatively treated with high-dose methotrexate, doxorubicin, and cisplatin. J. Clin. Oncol. 1998, 16, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.R.; Papadopolous, N.; Raymond, A.K.; Donato, M.; Seong, C.M.; Yasko, A.W.; Lewis, V.O.; Lin, P.P.; Champlin, R.; Benjamin, R.S. A phase II study of cisplatin, doxorubicin, and ifosfamide with peripheral blood stem cell support in patients with skeletal osteosarcoma and variant bone tumors with a poor prognosis. Cancer 2004, 101, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Minotti, G.; Menna, P.; Salvatorelli, E.; Cairo, G.; Gianni, L. Anthracyclines: Molecular Advances and Pharmacologic Developments in Antitumor Activity and Cardiotoxicity. Pharmacol. Rev. 2004, 56, 185–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owen, R.; Giacosa, A.; Hull, W.E.; Haubner, R.; Würtele, G.; Spiegelhalder, B.; Bartsch, H. Olive-oil consumption and health: The possible role of antioxidants. Lancet Oncol. 2000, 1, 107–112. [Google Scholar] [CrossRef]
- Zoidou, E.; Melliou, E.; Gikas, E.; Tsarbopoulos, A.; Magiatis, P.; Skaltsounis, A.-L. Identification of Throuba Thassos, a Traditional Greek Table Olive Variety, as a Nutritional Rich Source of Oleuropein. J. Agric. Food Chem. 2010, 58, 46–50. [Google Scholar] [CrossRef]
- Bonoli, M.; Bendini, A.; Cerretani, L.; Lercker, G.; Toschi, T.G. Qualitative and Semiquantitative Analysis of Phenolic Compounds in Extra Virgin Olive Oils as a Function of the Ripening Degree of Olive Fruits by Different Analytical Techniques. J. Agric. Food Chem. 2004, 52, 7026–7032. [Google Scholar] [CrossRef]
- Tripoli, E.; Giammanco, M.; Tabacchi, G.; Di Majo, D.; Giammanco, S.; La Guardia, M. The phenolic compounds of olive oil: Structure, biological activity and beneficial effects on human health. Nutr. Res. Rev. 2005, 18, 98–112. [Google Scholar] [CrossRef]
- Petkov, V.; Manolov, P. Pharmacological analysis of the iridoid oleuropein. Arzneimittelforschung 1972, 22, 1476–1486. [Google Scholar]
- Hamdi, H.K.; Castellon, R. Oleuropein, a non-toxic olive iridoid, is an anti-tumor agent and cytoskeleton disruptor. Biochem. Biophys. Res. Commun. 2005, 334, 769–778. [Google Scholar] [CrossRef]
- Anter, J.; Fernández-Bedmar, Z.; Villatoro-Pulido, M.; Demyda-Peyrás, S.; Moreno-Millán, M.; Alonso-Moraga, Á; Muñoz-Serrano, A.; De Castro, M.D.L. A pilot study on the DNA-protective, cytotoxic, and apoptosis-inducing properties of olive-leaf extracts. Mutat. Res. Toxicol. Environ. Mutagen. 2011, 723, 165–170. [Google Scholar] [CrossRef]
- Andreadou, I.; Iliodromitis, E.K.; Mikros, E.; Constantinou, M.; Agalias, A.; Magiatis, P.; Skaltsounis, A.L.; Kamber, E.; Tsantili-Kakoulidou, A.; Kremastinos, D.T. The Olive Constituent Oleuropein Exhibits Anti-Ischemic, Antioxidative, and Hypolipidemic Effects in Anesthetized Rabbits. J. Nutr. 2006, 136, 2213–2219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruzzolini, J.; Peppicelli, S.; Andreucci, E.; Bianchini, F.; Scardigli, A.; Romani, A.; La Marca, G.; Nediani, C.; Calorini, L. Oleuropein, the Main Polyphenol of Olea europaea Leaf Extract, Has an Anti-Cancer Effect on Human BRAF Melanoma Cells and Potentiates the Cytotoxicity of Current Chemotherapies. Nutrients 2018, 10, 1950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherif, I.O.; Al-Gayyar, M.M. Oleuropein potentiates anti-tumor activity of cisplatin against HepG2 through affecting proNGF/NGF balance. Life Sci. 2018, 198, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Papachristodoulou, A.; Tsoukala, M.; Benaki, D.; Kostidis, S.; Gioti, K.; Aligiannis, N.; Pratsinis, H.; Kletsas, D.; Skaltsounis, A.-L.; Mikros, E.; et al. Oleuropein is a Powerful Sensitizer of Doxorubicin-mediated Killing of Prostate Cancer Cells and Exerts Its Action via Induction of Autophagy. J. Cancer Res. Treat. 2018, 4, 61–68. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.W.J.; Zhang, Q.; Li, X.; Zhu, X.; Wang, Q.; Cao, S.; Du, L. Mitochondria-mediated apoptosis was induced by oleuropein in H1299 cells involving activation of p38 MAP kinase. J. Cell Biochem. 2019, 120, 5480–5494. [Google Scholar] [CrossRef]
- Zhang, F.Z.M. Oleuropein inhibits esophageal cancer through hypoxic suppression of BTG3 mRNA. Food Funct. 2019, 10, 978–985. [Google Scholar] [CrossRef]
- Mizushima, N. Autophagy: Process and function. Genes Dev. 2007, 21, 2861–2873. [Google Scholar] [CrossRef] [Green Version]
- Boka, V.I.; Argyropoulou, A.; Gikas, E.; Angelis, A.; Aligiannis, N.; Skaltsounis, A.L. Employment of High-Performance Thin-Layer Chromatography for the Quantification of Oleuropein in Olive Leaves and the Selection of a Suitable Solvent System for Its Isolation with Centrifugal Partition Chromatography. Planta Med. 2015, 81, 1628–1635. [Google Scholar] [CrossRef] [Green Version]
- Vistica, D.T.; Skehan, P.; Scudiero, D.; Monks, A.; Pittman, A.; Boyd, M.R. Tetrazolium-based assays for cellular viability: A critical examination of selected parameters affecting formazan production. Cancer Res. 1991, 51, 2515–2520. [Google Scholar]
- Gioti, K.; Papachristodoulou, A.; Benaki, D.; Havaki, S.; Beloukas, A.; Vontzalidou, A.; Aligiannis, N.; Skaltsounis, A.L.; Mikros, E.; Tenta, R. Silymarin Enriched Extract (Silybum marianum) Additive Effect on Doxorubicin-Mediated Cytotoxicity in PC-3 Prostate Cancer Cells. Planta Med. 2019, 85, 997–1007. [Google Scholar] [CrossRef]
- Kowalik, M.A.; Perra, A.; Ledda-Columbano, G.M.; Lapadula, G.; Piacentini, M.; Columbano, A.; Falasca, L. Induction of autophagy promotes the growth of early preneoplastic rat liver nodules. Oncotarget 2015, 7, 5788–5799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toepfer, N.; Childress, C.; Parikh, A.; Rukstalis, D.; Yang, W. Atorvastatin induces autophagy in prostate cancer PC3 cells through activation ofLC3transcription. Cancer Biol. Ther. 2011, 12, 691–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Belle, J.E.; Harris, N.G.; Williams, S.R.; Bhakoo, K.K. A comparison of cell and tissue extraction techniques using high-resolution 1H-NMR spectroscopy. NMR Biomed. 2002, 15, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Duarte, I.F.; Lamego, I.; Marques, J.; Marques, M.P.M.; Blaise, B.J.; Gil, A.M. Nuclear Magnetic Resonance (NMR) Study of the Effect of Cisplatin on the Metabolic Profile of MG-63 Osteosarcoma Cells. J. Proteome Res. 2010, 9, 5877–5886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamego, I.; Duarte, I.F.; Marques, M.P.M.; Gil, A.M. Metabolic Markers of MG-63 Osteosarcoma Cell Line Response to Doxorubicin and Methotrexate Treatment: Comparison to Cisplatin. J. Proteome Res. 2014, 13, 6033–6045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choupani, J.A.M.; Derakhshan, S.M.; Zaeifizadeh, M.; Khaniani, M.S. Oleuropein inhibits migration ability through suppression of epithelial-mesenchymal transition and synergistically enhances doxorubicin-mediated apoptosis in MCF-7 cells. J. Cell Physiol. 2019, 234, 9093–9104. [Google Scholar] [CrossRef]
- Cai, F.; Luis, M.A.F.; Lin, X.; Wang, M.; Cai, L.; Cen, C.; Biskup, E. Anthracycline-induced cardiotoxicity in the chemotherapy treatment of breast cancer: Preventive strategies and treatment (Review). Mol. Clin. Oncol. 2019, 11, 15–23. [Google Scholar] [CrossRef]
- Capelôa, T.B.Z.; Zampieri, L.X.; Blackman, M.C.N.M.; Sonveaux, P. Metabolic and non-metabolic pathways that control cancer resistance to anthracyclines. Semin. Cell Dev. Biol. 2020, 98, 181–191. [Google Scholar] [CrossRef]
- Moran, J.M.; Leal-Hernandez, O.; Canal-Macías, M.L.; Roncero-Martin, R.; Guerrero-Bonmatty, R.; Aliaga, I.; Zamorano, J.D.P. Antiproliferative Properties of Oleuropein in Human Osteosarcoma Cells. Nat. Prod. Commun. 2016, 11, 491–492. [Google Scholar] [CrossRef] [Green Version]
- Elamin, M.H.; Elmahi, A.B.; Daghestani, M.; Al-Olayan, E.M.A.; Al-Ajmi, R.; Alkhuriji, A.F.; Hamed, S.S.; El-Khadragy, M.F. Synergistic Anti-Breast-Cancer Effects of Combined Treatment With Oleuropein and Doxorubicin In Vivo. Altern. Ther. Heal. Med. 2017, 25, 17–24. [Google Scholar]
- Bilim, V.; Kawasaki, T.; Takahashi, K.; Tomita, Y. Adriamycin induced G2/M cell cycle arrest in transitional cell cancer cells with wt p53 and p21(WAF1/CIP1) genes. J. Exp. Clin. Cancer Res. 2000, 19, 483–488. [Google Scholar] [PubMed]
- Ji, P.; Yu, L.; Guo, W.; Mei, H.-J.; Wang, X.-J.; Chen, H.; Fang, S.; Yang, J. Doxorubicin Inhibits Proliferation of Osteosarcoma Cells Through Upregulation of the Notch Signaling Pathway. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2015, 22, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Bell, D.F.; Bell, R.S.; Mankin, H.J.; Gebhardt, M.C.; Weltie, F.; O’Brien, T. Kinetic effects of adriamycin and bleomycin on two osteosarcoma models. J. Orthop. Res. 1988, 6, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Vella, S.; Tavanti, E.; Hattinger, C.M.; Fanelli, M.; Versteeg, R.; Koster, J.; Picci, P.; Serra, M. Targeting CDKs with Roscovitine Increases Sensitivity to DNA Damaging Drugs of Human Osteosarcoma Cells. PLoS ONE 2016, 11, e0166233. [Google Scholar] [CrossRef]
- White, E. The role for autophagy in cancer. J. Clin. Investig. 2015, 125, 42–46. [Google Scholar] [CrossRef] [Green Version]
- Mizushima, N.; Yoshimori, T. How to Interpret LC3 Immunoblotting. Autophagy 2007, 3, 542–545. [Google Scholar] [CrossRef]
- Wang, L.; Tang, B.; Han, H.; Mao, D.; Chen, J.; Zeng, Y.; Xiong, M. miR-155 Affects Osteosarcoma MG-63 Cell Autophagy Induced by Adriamycin Through Regulating PTEN-PI3K/AKT/mTOR Signaling Pathway. Cancer Biotherapy Radiopharm. 2018, 33, 32–38. [Google Scholar] [CrossRef]
- Cordero, J.G.; García-Escudero, R.; Avila, J.; Gargini, R.; García-Escudero, V. Benefit of Oleuropein Aglycone for Alzheimer’s Disease by Promoting Autophagy. Oxidative Med. Cell. Longev. 2018, 2018, 1–12. [Google Scholar] [CrossRef]
- Liu, M.; Wang, J.; Huang, B.; Chen, A.; Li, X. Oleuropein inhibits the proliferation and invasion of glioma cells via suppression of the AKT signaling pathway. Oncol. Rep. 2016, 36, 2009–2016. [Google Scholar] [CrossRef]
- Rigacci, S.; Miceli, C.; Nediani, C.; Berti, A.; Cascella, R.; Pantano, D.; Nardiello, P.; Luccarini, I.; Casamenti, F.; Stefania, R. Oleuropein aglycone induces autophagy via the AMPK/mTOR signalling pathway: A mechanistic insight. Oncotarget 2015, 6, 35344–35357. [Google Scholar] [CrossRef] [Green Version]
- Johansen, T.; Lamark, T. Selective autophagy mediated by autophagic adapter proteins. Autophagy 2011, 7, 279–296. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Arozena, A.A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puissant, A.; Auberger, P. AMPK- and p62/SQSTM1-dependent autophagy mediate Resveratrol-induced cell death in chronic myelogenous leukemia. Autophagy 2010, 6, 655–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawaguchi, M.A.S.; Hirao, T.; Morita, M.; Ito, K. Autophagy is an important metabolic pathway to determine leukemia cell survival following suppression of the glycolytic pathway. Biochem. Biophys. Res. Commun. 2016, 474, 188–192. [Google Scholar] [CrossRef] [PubMed]
- Duran, R.V.; Oppliger, W.; Robitaille, A.M.; Heiserich, L.; Skendaj, R.; Gottlieb, E.; Hall, M.N. Glutaminolysis activates Rag-mTORC1 signaling. Mol. Cell 2012, 47, 349–358. [Google Scholar] [CrossRef] [Green Version]
- Glunde, K.; Penet, M.-F.; Jiang, L.; Jacobs, M.A.; Bhujwalla, Z.M. Choline metabolism-based molecular diagnosis of cancer: An update. Expert Rev. Mol. Diagn. 2015, 15, 735–747. [Google Scholar] [CrossRef]
- Li, Z.; Wu, G.; Van Der Veen, J.N.; Hermansson, M.; Vance, D.E. Phosphatidylcholine metabolism and choline kinase in human osteoblasts. Biochim. et Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2014, 1841, 859–867. [Google Scholar] [CrossRef]
- Andrejeva, G.; Gowan, S.; Lin, G.; Fong, A.-C.L.W.T.; Shamsaei, E.; Parkes, H.G.; Mui, J.; Raynaud, F.I.; Asad, Y.; Vizcay-Barrena, G.; et al. De novo phosphatidylcholine synthesis is required for autophagosome membrane formation and maintenance during autophagy. Autophagy 2020, 16, 1044–1060. [Google Scholar] [CrossRef] [Green Version]
- Kreuzaler, P.; Panina, Y.; Segal, J.; Yuneva, M. Adapt and conquer: Metabolic flexibility in cancer growth, invasion and evasion. Mol. Metab. 2020, 33, 83–101. [Google Scholar] [CrossRef]
- Lu, H.-Y.; Zhu, J.-S.; Xie, J.; Zhang, Z.; Zhu, J.; Jiang, S.; Shen, W.-J.; Wu, B.; Ding, T.; Wang, S.-L. Hydroxytyrosol and Oleuropein Inhibit Migration and Invasion via Induction of Autophagy in ER-Positive Breast Cancer Cell Lines (MCF7 and T47D). Nutr. Cancer 2020, 14, 1–11. [Google Scholar] [CrossRef]
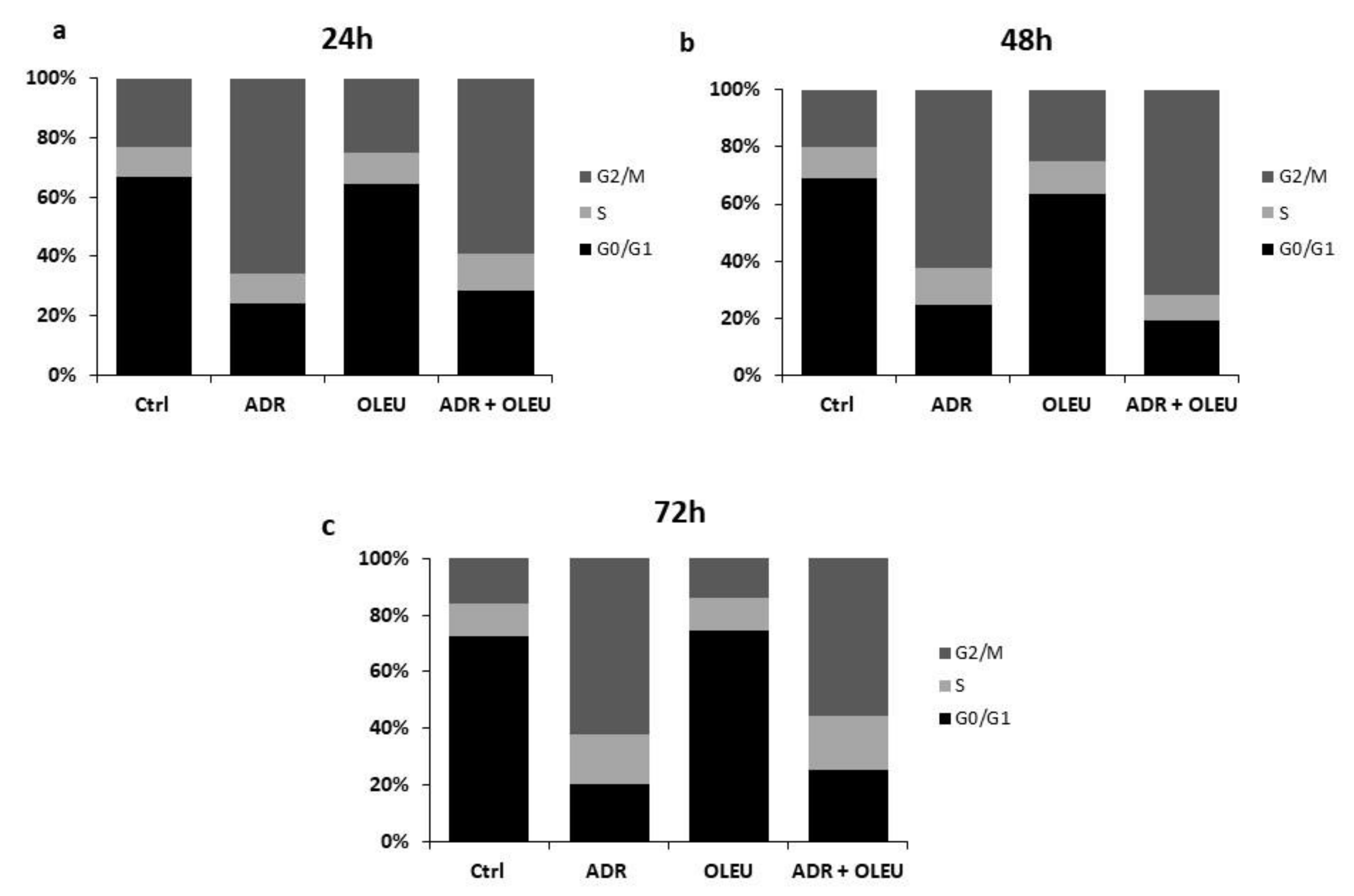

| Cell Cycle Phase Distribution | G0/G1 | S | G2/Μ | |
|---|---|---|---|---|
| Treatments | ||||
| 24 h | ||||
| Ctrl | 66.9 | 10.2 | 22.9 | |
| ADR | 24 | 10.1 | 65.9 | |
| OLEU | 64.6 | 10.5 | 24.9 | |
| ADR + OLEU | 28.3 | 12.6 | 59.1 | |
| 48 h | ||||
| Ctrl | 69 | 11.2 | 19.8 | |
| ADR | 24.6 | 12.8 | 62.6 | |
| OLEU | 63.3 | 11.9 | 24.8 | |
| ADR + OLEU | 19.1 | 9.2 | 71.7 | |
| 72 h | ||||
| Ctrl | 72.3 | 11.7 | 16 | |
| ADR | 20.1 | 17.7 | 62.2 | |
| OLEU | 74.6 | 11.4 | 14 | |
| ADR + OLEU | 25.4 | 19.2 | 55.4 | |
| No | Metabolites | Chemical Shift (ppm) (Multiplicity) |
|---|---|---|
| Amino Acids and Derivatives | ||
| 1 | Alanine (Ala) | 3.79 (q); 1.48 (d) |
| 2 | Arginine (Arg) | 3.25 (t); 1.94 (m) |
| 3 | Aspartate (Asp) | 2.81 (dd); 2.69 (dd) |
| 4 | Glutamate (Glu) | 3.76 (q); 2.35 (m); 2.13 (m); 2.06 (m) |
| 5 | Glycine (Gly) | 3.56 (s) |
| 6 | Histidine (His) | 7.87; 7.10 |
| 7 | Isoleucine (Ile) | 1.01 (d); 0.94 (t) |
| 8 | Leucine (Leu) | 0.97 (d); 0.96 (d) |
| 9 | Lysine (Lys) | 3.03 (t); 1.74 (m) |
| 10 | Phenylalanine (Phe) | 7.43 (m); 7.38 (m); 7.34 (d) |
| 11 | Threonine (Thr) | 1.335 (d) |
| 12 | Tyrosine (Tyr) | 7.20 (d); 6.90 (d) |
| 13 | Valine (Val) | 1.05 (d); 1.00 (d) |
| 14 | Creatine | 3.93 (s); 3.04 (s) |
| 15 | Creatine Phosphate | 3.045 (s) |
| 16 | Glutathione reduced (GSH) | 3.77 (m); 2.56 (m); 2.17 (m) |
| 17 | N-Acetylated Amino Acids (NAcAA) | 2.02 (s) |
| 18 | Taurine | 3.42 (t); 3.25 (t) |
| Organic Acids | ||
| 19 | Acetate | 1.92 (s) |
| 20 | Formate (Form) | 8.46 (s) |
| 21 | Lactate (Lac) | 4.11 (q); 1.33 (d) |
| 22 | Succinate (Succ) | 2.41 (s) |
| Nucleotides and Derivatives | ||
| 23 | Adenosine diphosphate (ADP) | 8.535 (s) |
| 24 | Adenosine triphosphate (ATP) | 8.545 (s); 8.28 (s); 6.15 (d); 4.62 (m); 4.41 (t) |
| 25 | Uridine Diphosphates (UDPs) | 7.98 (d); 5.98 (m) |
| 26 | Uridine Diphosphate (UDP-GlucNAc) | 7.95 (d); 5.98 (m); 2.08 (s) |
| Choline and Derivatives | ||
| 27 | Choline (Cho) | 3.21 (s) |
| 28 | O-Phosphocholine (PC) | 3.225 (s) |
| 29 | Glycerophosphocholine (GPC) | 3.235 (s) |
| Lipid Fatty Acids | ||
| 30 | CH3 | 0.88 |
| 31 | (CH2)n | 1.29 |
| 32 | CH2CH2CO | 1.55 |
| Other | ||
| 33 | Glucose (Glc) | 5.24 (d); 4.65 (d); 3.90 (dd); 3.84 (m); 3.73 (m); 3.50 (t); 3.47 (m); 3.41 (m); 3.25 (dd) |
| 34 | Myo-Inositol | 4.07 (t); 3.63 (t); 3.54 (dd); 3.29 (t) |
| 35 | Glycerol | 3.65 (dd); 3.56 (dd) |
| 36 | Methanol (Meth) | 3.36 (s) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gioti, K.; Papachristodoulou, A.; Benaki, D.; Aligiannis, N.; Skaltsounis, A.-L.; Mikros, E.; Tenta, R. Assessment of the Nutraceutical Effects of Oleuropein and the Cytotoxic Effects of Adriamycin, When Administered Alone and in Combination, in MG-63 Human Osteosarcoma Cells. Nutrients 2021, 13, 354. https://doi.org/10.3390/nu13020354
Gioti K, Papachristodoulou A, Benaki D, Aligiannis N, Skaltsounis A-L, Mikros E, Tenta R. Assessment of the Nutraceutical Effects of Oleuropein and the Cytotoxic Effects of Adriamycin, When Administered Alone and in Combination, in MG-63 Human Osteosarcoma Cells. Nutrients. 2021; 13(2):354. https://doi.org/10.3390/nu13020354
Chicago/Turabian StyleGioti, Katerina, Anastasia Papachristodoulou, Dimitra Benaki, Nektarios Aligiannis, Alexios-Leandros Skaltsounis, Emmanuel Mikros, and Roxane Tenta. 2021. "Assessment of the Nutraceutical Effects of Oleuropein and the Cytotoxic Effects of Adriamycin, When Administered Alone and in Combination, in MG-63 Human Osteosarcoma Cells" Nutrients 13, no. 2: 354. https://doi.org/10.3390/nu13020354








