The Changes in Endogenous Metabolites in Hyperlipidemic Rats Treated with Herbal Mixture Containing Lemon, Apple Cider, Garlic, Ginger, and Honey
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Sample Preparation
2.3. Ascorbic Acid (Vitamin C) Determination
2.4. Phenolic Extraction Using Solid-Phase Extraction (SPE) and Liquid-Liquid Extraction (LLE)
2.5. High-Performance Liquid Chromatography (HPLC) Analysis
2.6. Liquid Chromatography-Mass Spectroscopy (LC-MS)
2.7. Efficacy Study on an Animal Model
2.7.1. Acclimatization
2.7.2. Hyperlipidemia Induction
2.7.3. Treatment with the Polyphenol-Rich Mixture (PRM) and Simvastatin
- Group C: Normal/Control diet group, given 1 mL of filtered water
- Group HC: High cholesterol diet group, given 1 mL of filtered water
- Group PRM-H: High cholesterol diet group, given 500 mg/kg PRM
- Group PRM-M: High cholesterol diet group, given 250 mg/kg PRM
- Group PRM-L: High cholesterol diet group, given 150 mg/kg PRM
- Group D: High cholesterol diet group, given simvastatin 10 mg/kg
2.7.4. Blood and Urine Collection
2.7.5. Serum Biochemistry Analyses and HMG-CoA Reductase
2.8. 1H NMR Measurement, Data Processing and Multivariate Analysis
2.9. Statistical Analysis (Multivariate and Univariate Analysis)
3. Results
3.1. Characterization of PRM
3.2. Serum Biochemistry and Inhibition of HMG-CoA Reductase Activity
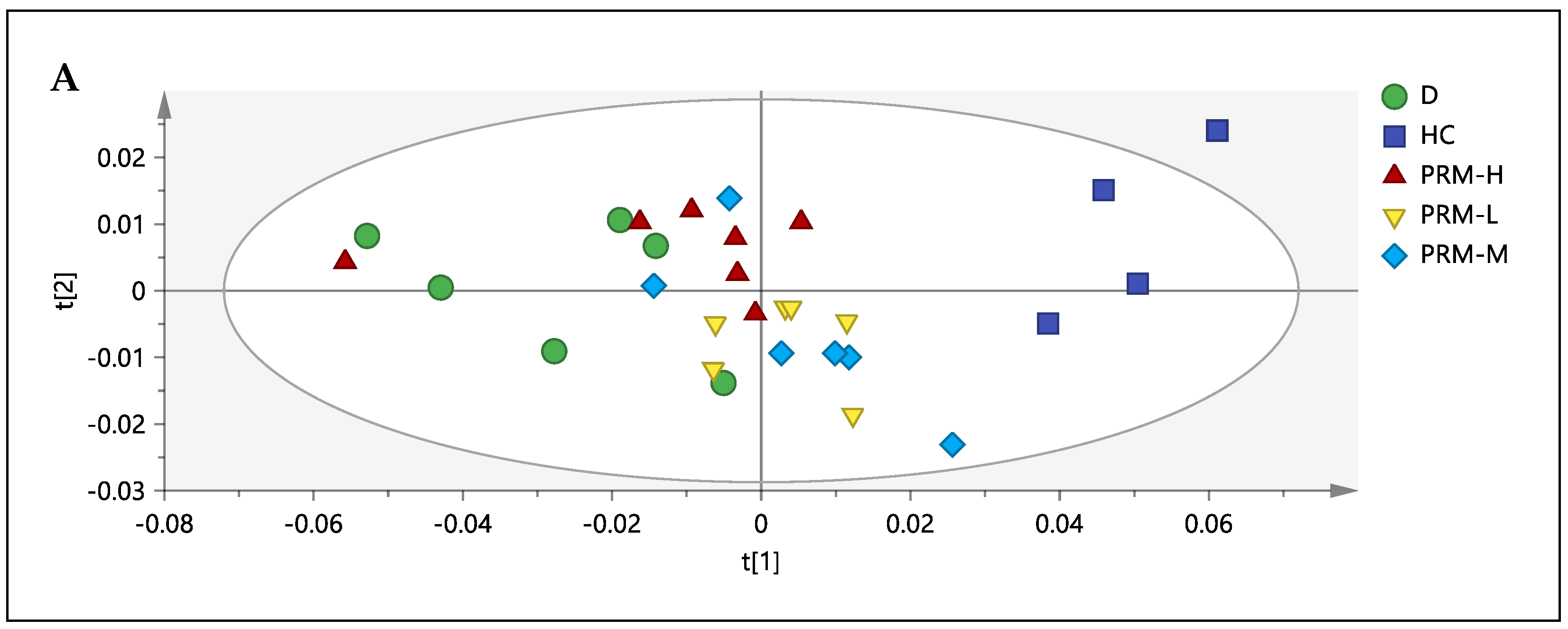
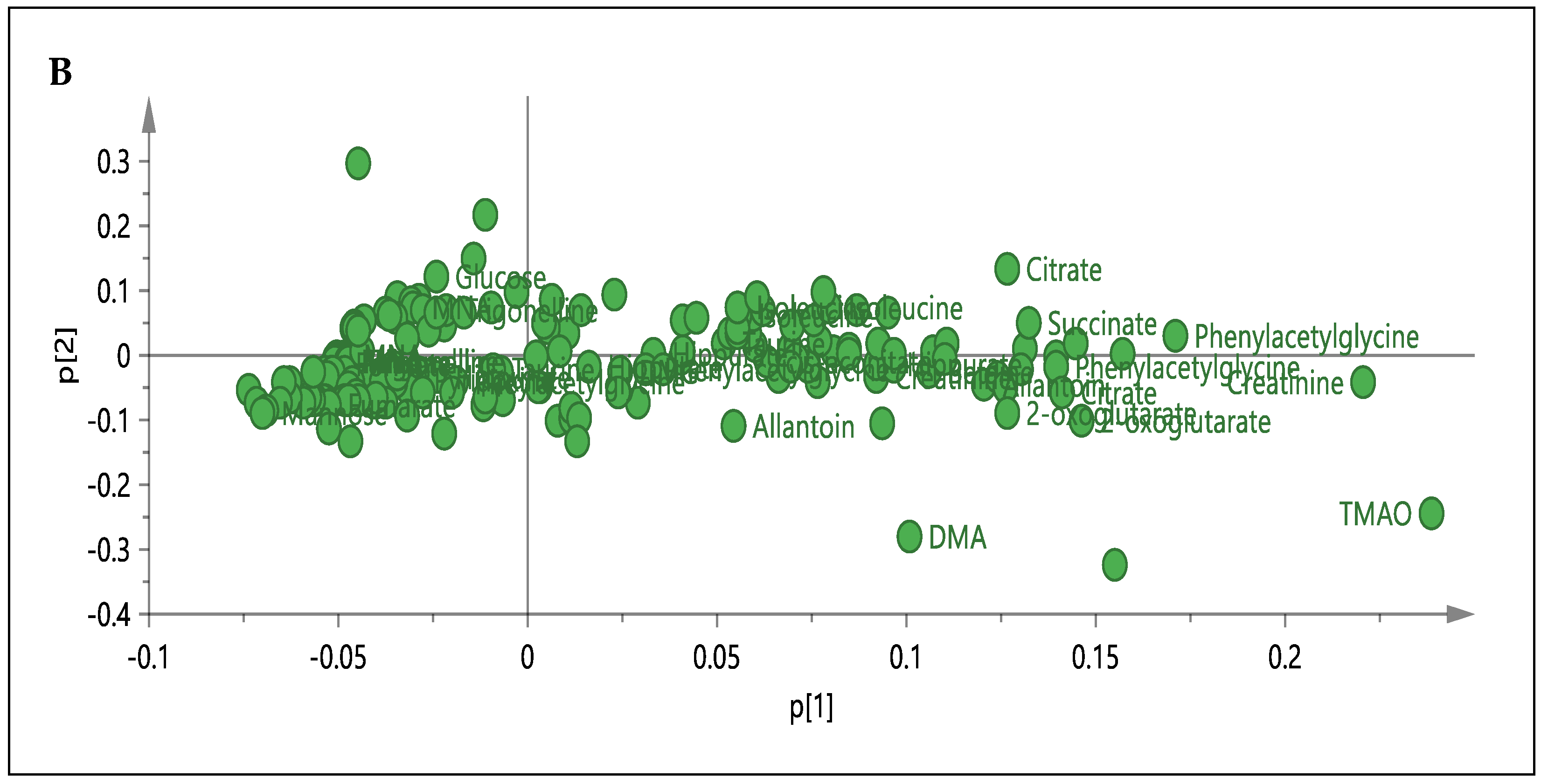
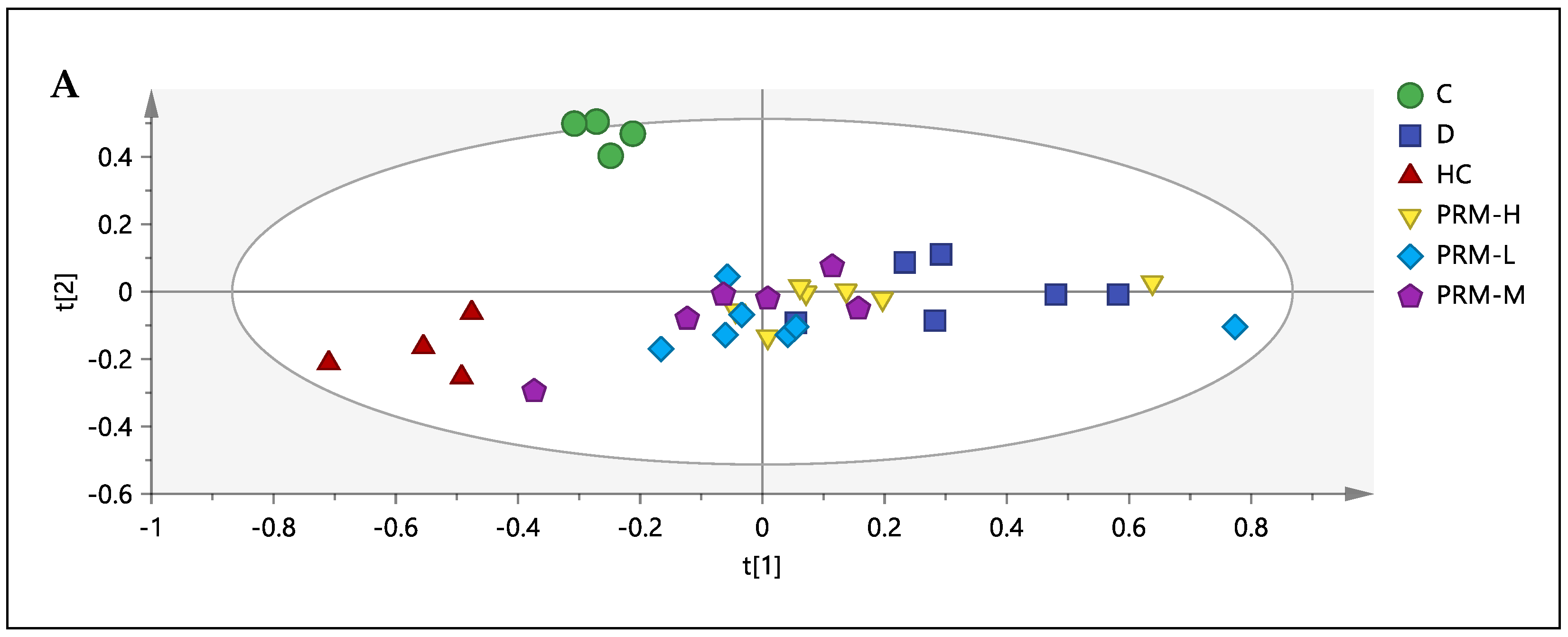
3.3. 1H-NMR Spectral Analysis of Urine Samples
3.4. Correlation between the Metabolites, HMG-CoA Reductase Activity and Low-Density Lipoprotein
3.5. Relative Quantification of Metabolites and Pathway Analysis
4. Discussion
4.1. Carbohydrate Metabolism
4.2. Amino Acid Metabolism
4.3. Gut Microbiota and the Other Metabolism
4.4. Study Limitations and Future Recommendations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Song, X.; Wang, J.; Wang, P.; Tian, N.; Yang, M.; Kong, L. 1H NMR-based metabolomics approach to evaluate the effect of Xue-Fu-Zhu-Yu decoction on hyperlipidemia rats induced by a high-fat diet. J. Pharm. Biomed. Anal. 2013, 78–79, 202–210. [Google Scholar] [CrossRef]
- Nelson, R.H. Hyperlipidemia as a risk factor for cardiovascular disease. Prim. Care 2013, 40, 195–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Noncommunicable Diseases. Available online: https://www.who.int/data/gho/data/themes/noncommunicable-diseases (accessed on 27 November 2020).
- Bełtowski, J.; Wójcicka, G.; Jamroz-Wiśniewska, A. Adverse effects of statins—mechanisms and consequences. Curr. Drug Saf. 2009, 4, 209–228. [Google Scholar] [CrossRef]
- Banach, M.; Patti, A.M.; Giglio, R.V.; Cicero, A.F.G.; Atanasov, A.G.; Bajraktari, G.; Bruckert, E.; Descamps, O.; Djuric, D.M.; Ezhov, M.; et al. The Role of Nutraceuticals in Statin Intolerant Patients. J. Am. Coll. Cardiol. 2018, 72, 96–118. [Google Scholar] [CrossRef]
- Chien, M.Y.; Ku, Y.H.; Chang, J.M.; Yang, C.M.; Chen, C.H. Effects of herbal mixture extracts on obesity in rats fed a high-fat diet. J. Food Drug Anal. 2016, 24, 594–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.-H.; Park, K.-S. Effects of Panax ginseng extract on lipid metabolism in humans. Pharmacol. Res. 2003, 48, 511–513. [Google Scholar] [CrossRef]
- He, Z.; Lei, L.; Kwek, E.; Zhao, Y.; Liu, J.; Hao, W.; Zhu, H.; Liang, N.; Ma, K.Y.; Ho, H.M.; et al. Ginger attenuates trimethylamine-N-oxide (TMAO)-exacerbated disturbance in cholesterol metabolism and vascular inflammation. J. Funct. Foods 2019, 52, 25–33. [Google Scholar] [CrossRef]
- Alizadeh-Navaei, R.; Roozbeh, F.; Saravi, M.; Pouramir, M.; Jalali, F.; Moghadamnia, A.A. Investigation of the effect of ginger on the lipid levels. A double-blind controlled clinical trial. Saudi Med. J. 2008, 29, 1280–1284. [Google Scholar] [PubMed]
- Panahi, Y.; Ahmadi, Y.; Teymouri, M.; Johnston, T.P.; Sahebkar, A. Curcumin as a potential candidate for treating hyperlipidemia: A review of cellular and metabolic mechanisms. J. Cell Physiol. 2018, 233, 141–152. [Google Scholar] [CrossRef]
- Bencheikh, N.; Bouhrim, M.; Merrouni, I.A.; Boutahiri, S.; Kharchoufa, L.; Addi, M.; Tungmunnithum, D.; Hano, C.; Eto, B.; Legssyer, A.; et al. Antihyperlipidemic and antioxidant activities of flavonoid-rich extract of Ziziphus lotus (L.) lam. fruits. Appl. Sci. 2021, 11, 7788. [Google Scholar] [CrossRef]
- Ji, X.; Shi, S.; Liu, B.; Shan, M.; Tang, D.; Zhang, W.; Zhang, Y.; Zhang, L.; Zhang, H.; Lu, C.; et al. Bioactive compounds from herbal medicines to manage dyslipidemia. Biomed. Pharmacother. 2019, 118, 109338. [Google Scholar] [CrossRef]
- Firenzuoli, F.; Gori, L. Herbal medicine today: Clinical and research issues. Evid. Based Complement. Altern. Med. 2007, 4, 37–40. [Google Scholar] [CrossRef]
- Fürst, R.; Zündorf, I. Evidence-Based Phytotherapy in Europe: Where Do We Stand? Planta Med. 2015, 81, 962–967. [Google Scholar] [CrossRef]
- Naseem, E.; Shamim, M.; Khan, N.I. Cardioprotective effects of herbal mixture (ginger, garlic, lemon, apple cider vinegar and honey) in experimental animal models of hyperlipidemia. Int. J. Biol. Res. 2016, 4, 28–33. [Google Scholar]
- Naseem, A.; Akhtar, S.; Manzoor, M.F.; Sameen, A.; Layla, A.; Afzal, K.; Karrar, E.; Rahaman, A.; Ismail, T.; Ahmad, N.; et al. Effect of herbal formulation intake on health indices in albino Wistar rat model. Food Sci. Nutr. 2020, 9, 441–448. [Google Scholar] [CrossRef]
- Ishak, I.; George, P.; Ibrahim, F.W.; Yahya, H.M.; Farah, N. Acute modulatory effects of apple cider vinegar, garlic, ginger, lemon and honey mixture, with and without exercise on postprandial glycemia in non-diabetic females. J. Sains Kesihat. Malays. Isu Khas 2018, 16, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Sajak, A.A.B.; Mediani, A.; Dom, N.S.M.; Machap, C.; Hamid, M.; Ismail, A.; Khatib, A.; Abas, F. Effect of Ipomoea aquatica ethanolic extract in streptozotocin (STZ) induced diabetic rats via 1H NMR-based metabolomics approach. Phytomed 2017, 36, 201–209. [Google Scholar] [CrossRef]
- Wu, Z.; Mao, Y.; Zhang, X.; Weng, P. Symbolic metabolite analysis of pickled wax gourd in Eastern China by 1H-NMR spectroscopy and multivariate data. Int. J. Food Prop. 2016, 19, 2052–2062. [Google Scholar] [CrossRef]
- Diao, C.; Zhao, L.; Guan, M.; Zheng, Y.; Chen, M.; Yang, Y.; Lin, L.; Chen, W.; Gao, H. Systemic and characteristic metabolites in the serum of streptozotocin-induced diabetic rats at different stages as revealed by a 1H-NMR based metabonomic approach. Mol. Biosyst. 2014, 10, 686–693. [Google Scholar] [CrossRef]
- Abu Bakar Sajak, A.; Azlan, A.; Abas, F.; Hamzah, H. Nutritional composition, phytochemicals and acute toxicity of herbal mixture (lemon, apple cider, garlic, ginger and honey) in the zebrafish embryo and Wistar rat. Food Res. 2020, 4, 196–204. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis, 16th ed.; Method 967.21; AOAC International: Rockville, MD, USA, 1995. [Google Scholar]
- Jasicka-Misiak, I.; Makowicz, E.; Stanek, N. Polish yellow sweet clover (Melilotus officinalis L.) honey, chromatographic fingerprints, and chemical markers. Molecules 2017, 22, 138. [Google Scholar] [CrossRef] [Green Version]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Eriksson, L.; Johansson, E.; Kettaneh-Wold, N.; Wold, S.; Trygg, J.; Wikstrom, C. Multi- and Megavariate data Analysis Part I: Basic Principles and Applications, 2nd ed.; Umetrics Academy (Appendix I): Umea, Sweden, 2009. [Google Scholar]
- Jose, J. Statins and its hepatic effects: Newer data, implications, and changing recommendations. J. Pharm. Bioallied. Sci. 2016, 8, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Arafa, HM Curcumin attenuates diet-induced hypercholesterolemia in rats. Med. Sci. Monit. 2005, 11, BR228–BR234.
- Li, X.; Guo, J.; Liang, N.; Jiang, X.; Song, Y.; Ou, S.; Hu, Y.; Jiao, R.; Bai, W. 6-Gingerol regulates hepatic cholesterol metabolism by up-regulation of LDLR and cholesterol efflux-related genes in HepG2 cells. Front. Pharmacol. 2018, 9, 159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; McNeill, J.H. Ascorbic acid supplementation prevents hyperlipidemia and improves myocardial performance in streptozotocin-diabetic rats. Diabetes Res. Clin. Pract. 1995, 27, 11–18. [Google Scholar] [CrossRef]
- Lei, L.; Liu, Y.; Wang, X.; Jiao, R.; Ma, K.Y.; Li, Y.M.; Wang, L.; Man, S.W.; Sang, S.; Huang, Y.; et al. Plasma cholesterol-lowering activity of gingerol- and shogaol-enriched extract is mediated by increasing sterol excretion. J. Agric. Food Chem. 2014, 62, 10515–10521. [Google Scholar] [CrossRef]
- Wang, Y.K.; Hong, Y.J.; Yao, Y.H.; Huang, X.M.; Liu, X.B.; Zhang, C.Y.; Zhang, L.; Xu, X.L. 6-Shogaol protects against oxidized LDL-induced endothelial injuries by inhibiting oxidized LDL-evoked LOX-1 signaling. Evid. Based Complement. Altern. Med. 2013, 2013, 503521. [Google Scholar] [CrossRef]
- Nammi, S.; Kim, M.-S.; Gavande, N.; Li, G.; Roufogalis, B. Regulation of low-Density lipoprotein receptor and 3-hydroxy-3-methylglutaryl Coenzyme A reductase expression by Zingiber officinale in the liver of high-fat diet-fed rats. Basic Clin. Pharmacol. Toxicol. 2009, 106, 389–395. [Google Scholar] [CrossRef]
- Li, W.; Qu, X.-N.; Han, Y.; Zheng, S.-W.; Wang, J.; Wang, Y.-P. Ameliorative effects of 5-Hydroxymethyl-2-furfural (5-HMF) from Schisandra chinensis on alcoholic liver oxidative injury in mice. Int. J. Mol. Sci. 2015, 16, 2446–2457. [Google Scholar] [CrossRef]
- Jiang, C.Y.; Yang, K.M.; Yang, L.; Miao, Z.X.; Wang, Y.H.; Zhu, H.B. A 1H NMR-Based Metabonomic Investigation of Time-Related Metabolic Trajectories of the Plasma, Urine and Liver Extracts of Hyperlipidemic Hamsters. PLoS ONE 2013, 8, e66786. [Google Scholar] [CrossRef] [Green Version]
- Fraulob, J.C.; Souza-Mello, V.; Aguila, M.B.; Mandarim-de-Lacerda, C.A. Beneficial effects of rosuvastatin on insulin resistance, adiposity, inflammatory markers and non-alcoholic fatty liver disease in mice fed on a high-fat diet. Clin. Sci. 2012, 123, 259–270. [Google Scholar] [CrossRef] [Green Version]
- Han, K.H. Functional implications of HMG-CoA reductase inhibition on glucose metabolism. Korean Circ. J. 2018, 48, 951–963. [Google Scholar] [CrossRef]
- Bai, X.P.; Dong, F.; Yang, G.H.; Zhang, L. Influences of sterol regulatory element-binding protein-1c silencing on glucose production in HepG2 cells treated with free fatty acid. Lipids Health Dis. 2019, 18, 89. [Google Scholar] [CrossRef] [Green Version]
- Wurtz, P.; Soininen, P.; Kangas, A.J.; Ronnemaa, T.; Lehtimaki, T.; Kahonen, M.; Viikari, J.S.; Raitakari, O.T.; Ala-Korpela, M. Branched-chain and aromatic amino acids are predictors of insulin resistance in young adults. Diabetes Care 2013, 36, 648–655. [Google Scholar] [CrossRef] [Green Version]
- Siddik, M.A.B.; Shin, A.C. Recent progress on branched-chain amino acids in obesity, diabetes, and beyond. Endocrinol. Metab. 2019, 34, 234–246. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Hu, W.; Fu, Z.; Sun, L.; Zhou, Y.; Gong, Y.; Yang, T.; Zhou, H. The positive association of branched-chain amino acids and metabolic dyslipidemia in Chinese Han population. Lipids Health Dis. 2016, 15, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sookoian, S.; Pirola, C.J. Alanine and aspartate aminotransferase and glutamine-cycling pathway: Their roles in pathogenesis of metabolic syndrome. World J. Gastroenterol. 2012, 18, 3775–3781. [Google Scholar] [CrossRef]
- Filla, L.A.; Yuan, W.; Feldman, E.L.; Li, S.; Edwards, J.L. Global metabolomic and isobaric tagging capillary liquid chromatography-tandem mass spectrometry approaches for uncovering pathway dysfunction in diabetic mouse aorta. J. Proteome Res. 2014, 13, 6121–6134. [Google Scholar] [CrossRef]
- Fujisaka, S.; Avila-Pacheco, J.; Soto, M.; Kostic, A.; Dreyfuss, J.M.; Pan, H.; Ussar, S.; Altindis, E.; Li, N.; Bry, L.; et al. Diet, genetics, and the gut microbiome drive dynamic changes in plasma metabolites. Cell Rep. 2018, 22, 3027–3086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Wang, D.; Guo, S.; He, H.; Gong, L.; Cui, H. Gut microbiome and serum metabolome analyses identify unsaturated fatty acids and butanoate metabolism induced by gut microbiota in patients with chronic spontaneous urticaria. Front. Cell. Infect. Microbiol. 2020, 10, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pallister, T.; Jackson, M.A.; Martin, T.C.; Zierer, J.; Jennings, A.; Mohney, R.P.; MacGregor, A.; Steves, C.J.; Cassidy, A.; Spector, T.D.; et al. Hippurate as a metabolomic marker of gut microbiome diversity: Modulation by diet and relationship to metabolic syndrome. Sci. Rep. 2017, 7, 13670. [Google Scholar] [CrossRef] [Green Version]
- Anwar, S.; Bhandari, U.; Panda, B.P.; Dubey, K.; Khan, W.; Ahmad, S. Trigonelline inhibits intestinal microbial metabolism of choline and its associated cardiovascular risk. J. Pharm. Biomed. Anal. 2018, 159, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Sharma, L.; Lone, N.A.; Knott, R.M.; Hassan, A.; Abdullah, T. Trigonelline prevents high cholesterol and high-fat diet-induced hepatic lipid accumulation and lipo-toxicity in C57BL/6J mice via restoration of hepatic autophagy. Food Chem. Toxicol. 2018, 121, 283–296. [Google Scholar] [CrossRef]


| No | Sample | RT | Identified Compound | Weight (µg/500 mg of PRM) |
|---|---|---|---|---|
| 1 | PRM acidified | 4.84 | 5-Hydroxymethyfurfural (5-HMF) | 1126.77 ± 21.08 |
| 2 | Ethyl acetate fraction | 25.76 | 6-gingerol | 8.21 ± 0.44 |
| 2 | Ethyl acetate fraction | 31.26 | 6-shogaol | 45.06 ± 2.57 |
| Test | AST | ALT | Chol | Tgl | LDL-C | HDL-C | HMGCoA Reductase Activity (pg/mL) |
|---|---|---|---|---|---|---|---|
| Group | U/L | U/L | mmol/L | mmol/L | mmol/L | mmol/L | |
| Control (C) | 66.50 ± 45.96 | 78.50 ± 58.69 | 1.65 ± 0.21 # | 1.04 ± 0.03 | 0.16 ± 0.00 # | 1.46 ± 0.23 | 241.61 ± 44.48 # |
| Hyperlipidemia (HC) | 111.00 ± 28.87 | 144.00 ± 40.41 | 2.21 ± 0.19 | 0.82 ± 0.17 | 0.89 ± 0.11 | 1.16 ± 0.05 | 389.71 ± 64.92 * |
| PRM High Dose (PRM-H) | 106.75 ± 17.73 | 87.75 ± 29.92 | 1.55 ± 0.17 # | 0.76 ± 0.27 | 0.48 ± 0.10 * # | 1.21 ± 0.12 | 291.93 ± 20.19 # |
| PRM Medium Dose (PRM-M) | 138.60 ± 15.51 * | 61.80 ± 33.12 | 1.74 ± 0.17 # | 0.85 ± 0.16 | 0.56 ± 0.14 * # | 1.28 ± 0.11 | 309.61 ± 63.12 # |
| PRM Low Dose (PRM-L) | 126.50 ± 22.19 | 79.75 ± 18.28 | 1.85 ± 0.05 | 0.78 ± 0.24 | 0.64 ± 0.15 * | 1.31 ± 0.06 # | 372.81 ± 34.49 * |
| Simvastatin (D) | 148.50 ± 30.44 * | 85.00 ± 56.00 | 1.70 ± 0.25 # | 0.97 ± 0.21 | 0.64 ± 0.18 * | 1.15 ± 0.12 | 241.16 ± 56.60 # |
| Metabolites | δ 1H (ppm) | VIP | HC vs. C | PRM-H vs. HC | D vs. HC | |
|---|---|---|---|---|---|---|
| 1 | 2 | |||||
| Trimethylamine-N-oxide (TMAO) | 3.26 | 6.75 | 4.92 | H *** | L ** | L ** |
| Creatinine | 3.02 | 3.06 | 2.18 | L | L *** | L *** |
| Phenylacetylglycine | 3.66 | 1.71 | 2.89 | L | L ** | L *** |
| 2-oxoglutarate | 2.42, 2.98 | 0.39 | 2.88 | L ** | L ** | L ** |
| Succinate | 2.38 | 3.20 | 1.99 | H | L ** | L ** |
| Citrate | 2.66 | 1.20 | 3.67 | L | L * | L *** |
| Allantoin | 5.38 | 1.36 | 1.25 | H | L * | L *** |
| Dimethylamine (DMA) | 2.70 | 1.10 | 1.34 | L | L | L |
| Hippurate | 3.94 | 1.04 | 1.92 | L | L ** | L *** |
| Isoleucine | 1.22, 1.42 | 1.04 | 0.78 | H * | L ** | L ** |
| Cis-aconitate | 3.10 | 0.35 | 1.01 | L * | L ** | L * |
| Taurine | 3.25 | 0.86 | 1.14 | L | L | L * |
| Betaine | 3.89 | 1.56 | 1.06 | H | L ** | L ** |
| Fumarate | 6.51 | 0.61 | 0.55 | H | H *** | H *** |
| Formate | 8.44 | 0.31 | 0.74 | H | H ** | H |
| Dimethylglycine (DMG) | 2.91 | 0.76 | 0.60 | H * | L | L |
| Trigonelline | 9.10, 8.82 | 0.39 | 0.76 | L | H ** | H |
| Pyridoxine | 7.66 | 0.10 | 0.54 | L | H | H |
| Methylnicotinamide (MNA) | 9.26, 8.94, 8.86 | 0.42 | 0.86 | H | H ** | H ** |
| Mannose | 5.17 | 0.78 | 0.68 | H | H * | H * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abu Bakar Sajak, A.; Azlan, A.; Abas, F.; Hamzah, H. The Changes in Endogenous Metabolites in Hyperlipidemic Rats Treated with Herbal Mixture Containing Lemon, Apple Cider, Garlic, Ginger, and Honey. Nutrients 2021, 13, 3573. https://doi.org/10.3390/nu13103573
Abu Bakar Sajak A, Azlan A, Abas F, Hamzah H. The Changes in Endogenous Metabolites in Hyperlipidemic Rats Treated with Herbal Mixture Containing Lemon, Apple Cider, Garlic, Ginger, and Honey. Nutrients. 2021; 13(10):3573. https://doi.org/10.3390/nu13103573
Chicago/Turabian StyleAbu Bakar Sajak, Azliana, Azrina Azlan, Faridah Abas, and Hazilawati Hamzah. 2021. "The Changes in Endogenous Metabolites in Hyperlipidemic Rats Treated with Herbal Mixture Containing Lemon, Apple Cider, Garlic, Ginger, and Honey" Nutrients 13, no. 10: 3573. https://doi.org/10.3390/nu13103573