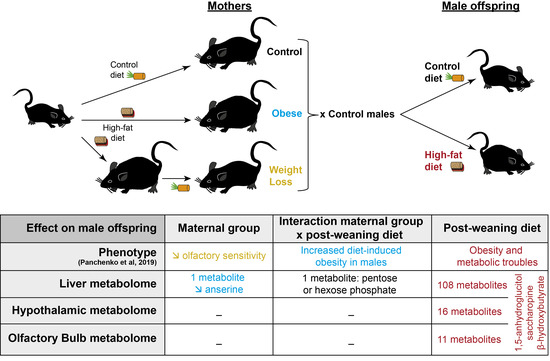
Differential Effects of Post-Weaning Diet and Maternal Obesity on Mouse Liver and Brain Metabolomes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Processing and Preparation
2.2. Liquid Chromatography–High-Resolution Mass Spectrometry (LC–HRMS) Metabolite Analyses
2.3. Biostatistical Analysis
2.4. Over-Representation Analysis
2.5. Pathway Analyses
3. Results
3.1. Nervous and Non-Nervous Tissues Display Similar Metabolite Profiles
3.2. Post-Weaning Diets Affect the Metabolite Profiles of both the Liver and Hypothalamus
3.3. Post-Weaning Diet Has a Major Impact on Metabolite Abundance in the Liver
3.4. Maternal Diet Affects the Liver Abundance of Two Metabolites
4. Discussion
4.1. The Liver, Hypothalamus and Olfactory Bulb Metabolomes Were Principally Affected by Chronic HFD
4.2. The Maternal Environment Has a Persistent Effect on Metabolites in the Liver
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organisation Obesity and Overweight. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 16 January 2020).
- Lee, C.Y.W.; Koren, G. Maternal obesity: Effects on pregnancy and the role of pre-conception counselling. J. Obstet. Gynaecol. J. Inst. Obstet. Gynaecol. 2010, 30, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Acosta, O.; Ramirez, V.I.; Lager, S.; Gaccioli, F.; Dudley, D.J.; Powell, T.L.; Jansson, T. Increased glucose and placental GLUT-1 in large infants of obese nondiabetic mothers. Am. J. Obstet. Gynecol. 2015, 212, 227-e1. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, L.; Ferraro, Z.M.; Wen, S.W.; Walker, M. Maternal obesity and occurrence of fetal macrosomia: A systematic review and meta-analysis. BioMed Res. Int. 2014, 2014, 640291. [Google Scholar] [CrossRef] [PubMed]
- Rajasingam, D.; Seed, P.T.; Briley, A.L.; Shennan, A.H.; Poston, L. A prospective study of pregnancy outcome and biomarkers of oxidative stress in nulliparous obese women. Am. J. Obstet. Gynecol. 2009, 200, 395-e1. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.; Seki, Y.; Vuguin, P.M.; Charron, M.J. Animal models of in utero exposure to a high fat diet: A review. Biochim. Biophys. Acta 2014, 1842, 507–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanson, M.A.; Gluckman, P.D. Early Developmental Conditioning of Later Health and Disease: Physiology or Pathophysiology? Physiol. Rev. 2014, 94, 1027–1076. [Google Scholar] [CrossRef]
- Hanson, M.; Barker, M.; Dodd, J.M.; Kumanyika, S.; Norris, S.; Steegers, E.; Stephenson, J.; Thangaratinam, S.; Yang, H. Interventions to prevent maternal obesity before conception, during pregnancy, and post partum. Lancet Diabetes Endocrinol. 2017, 5, 65–76. [Google Scholar] [CrossRef]
- Forsum, E.; Brantsæter, A.L.; Olafsdottir, A.-S.; Olsen, S.F.; Thorsdottir, I. Weight loss before conception: A systematic literature review. Food Nutr. Res. 2013, 57. [Google Scholar] [CrossRef]
- Dearden, L.; Bouret, S.G.; Ozanne, S.E. Sex and gender differences in developmental programming of metabolism. Mol. Metab. 2018, 15, 8–19. [Google Scholar] [CrossRef]
- McCurdy, C.E.; Bishop, J.M.; Williams, S.M.; Grayson, B.E.; Smith, M.S.; Friedman, J.E.; Grove, K.L. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J. Clin. Investig. 2009, 119, 323–335. [Google Scholar] [CrossRef] [Green Version]
- Del Mar Plata, M.; Williams, L.; Seki, Y.; Hartil, K.; Kaur, H.; Lin, C.-L.; Fiallo, A.; Glenn, A.S.; Katz, E.B.; Fuloria, M.; et al. Critical periods of increased fetal vulnerability to a maternal high fat diet. Reprod. Biol. Endocrinol. RBE 2014, 12, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lomas-Soria, C.; Reyes-Castro, L.A.; Rodríguez-González, G.L.; Ibáñez, C.A.; Bautista, C.J.; Cox, L.A.; Nathanielsz, P.W.; Zambrano, E. Maternal obesity has sex-dependent effects on insulin, glucose and lipid metabolism and the liver transcriptome in young adult rat offspring. J. Physiol. 2018, 596, 4611–4628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholas, L.M.; Rattanatray, L.; Morrison, J.L.; Kleemann, D.O.; Walker, S.K.; Zhang, S.; MacLaughlin, S.; McMillen, I.C. Maternal obesity or weight loss around conception impacts hepatic fatty acid metabolism in the offspring. Obes. Silver Spring Md 2014, 22, 1685–1693. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.-Q.; Gaysinskaya, V.; Karatayev, O.; Leibowitz, S.F. Maternal High-Fat Diet and Fetal Programming: Increased Proliferation of Hypothalamic Peptide-Producing Neurons That Increase Risk for Overeating and Obesity. J. Neurosci. 2008, 28, 12107–12119. [Google Scholar] [CrossRef] [Green Version]
- Sanders, T.R.; Kim, D.W.; Glendining, K.A.; Jasoni, C.L. Maternal Obesity and IL-6 Lead to Aberrant Developmental Gene Expression and Deregulated Neurite Growth in the Fetal Arcuate Nucleus. Endocrinology 2014, 155, 2566–2577. [Google Scholar] [CrossRef]
- Peleg-Raibstein, D.; Sarker, G.; Litwan, K.; Krämer, S.D.; Ametamey, S.M.; Schibli, R.; Wolfrum, C. Enhanced sensitivity to drugs of abuse and palatable foods following maternal overnutrition. Transl. Psychiatry 2016, 6, e911. [Google Scholar] [CrossRef]
- Sullivan, E.L.; Riper, K.M.; Lockard, R.; Valleau, J.C. Maternal high-fat diet programming of the neuroendocrine system and behavior. Horm. Behav. 2015, 76, 153–161. [Google Scholar] [CrossRef] [Green Version]
- Vogt, M.C.; Paeger, L.; Hess, S.; Steculorum, S.M.; Awazawa, M.; Hampel, B.; Neupert, S.; Nicholls, H.T.; Mauer, J.; Hausen, A.C.; et al. Neonatal Insulin Action Impairs Hypothalamic Neurocircuit Formation in Response to Maternal High-Fat Feeding. Cell 2014, 156, 495–509. [Google Scholar] [CrossRef] [Green Version]
- Merle, L.; Person, O.; Bonnet, P.; Grégoire, S.; Soubeyre, V.; Grosmaitre, X.; Jarriault, D. Maternal high fat high sugar diet disrupts olfactory behavior but not mucosa sensitivity in the offspring. Psychoneuroendocrinology 2019, 104, 249–258. [Google Scholar] [CrossRef] [Green Version]
- Panchenko, P.E.; Lacroix, M.-C.; Jouin, M.; Voisin, S.; Badonnel, K.; Lemaire, M.; Meunier, N.; Safi-Stibler, S.; Persuy, M.-A.; Jouneau, L.; et al. Effect of Maternal Obesity and Preconceptional Weight Loss on Male and Female Offspring Metabolism and Olfactory Performance in Mice. Nutrients 2019, 11, 948. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Li, X.; Niu, W.; Ma, G.; Sun, Q.; Bi, Y.; Guo, Z.; Ren, D.; Hu, J.; Yuan, F.; et al. Metabolomic profiling on rat brain of prenatal malnutrition: Implicated for oxidative stress and schizophrenia. Metab. Brain Dis. 2019, 34, 1607–1613. [Google Scholar] [CrossRef]
- Zhu, C.; Han, T.-L.; Zhao, Y.; Zhou, X.; Mao, X.; Qi, H.; Baker, P.N.; Zhang, H. A mouse model of pre-pregnancy maternal obesity combined with offspring exposure to a high-fat diet resulted in cognitive impairment in male offspring. Exp. Cell Res. 2018, 368, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Panchenko, P.E.; Voisin, S.; Jouin, M.; Jouneau, L.; Prézelin, A.; Lecoutre, S.; Breton, C.; Jammes, H.; Junien, C.; Gabory, A. Expression of epigenetic machinery genes is sensitive to maternal obesity and weight loss in relation to fetal growth in mice. Clin. Epigenetics 2016, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Hivert, M.F.; Perng, W.; Watkins, S.M.; Newgard, C.S.; Kenny, L.C.; Kristal, B.S.; Patti, M.E.; Isganaitis, E.; DeMeo, D.L.; Oken, E.; et al. Metabolomics in the developmental origins of obesity and its cardiometabolic consequences. J. Dev. Orig. Health Dis. 2015, 6, 65–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Hastings, J.; Owen, G.; Dekker, A.; Ennis, M.; Kale, N.; Muthukrishnan, V.; Turner, S.; Swainston, N.; Mendes, P.; Steinbeck, C. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016, 44, D1214–D1219. [Google Scholar] [CrossRef]
- Giacomoni, F.; Le Corguillé, G.; Monsoor, M.; Landi, M.; Pericard, P.; Pétéra, M.; Duperier, C.; Tremblay-Franco, M.; Martin, J.-F.; Jacob, D.; et al. Workflow4Metabolomics: A collaborative research infrastructure for computational metabolomics. Bioinforma. Oxf. Engl. 2015, 31, 1493–1495. [Google Scholar] [CrossRef] [Green Version]
- Guitton, Y.; Tremblay-Franco, M.; Le Corguillé, G.; Martin, J.-F.; Pétéra, M.; Roger-Mele, P.; Delabrière, A.; Goulitquer, S.; Monsoor, M.; Duperier, C.; et al. Create, run, share, publish, and reference your LC-MS, FIA-MS, GC-MS, and NMR data analysis workflows with the Workflow4Metabolomics 3.0 Galaxy online infrastructure for metabolomics. Int. J. Biochem. Cell Biol. 2017, 93, 89–101. [Google Scholar] [CrossRef]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Lenth, R.V. Least-Squares Means: The R Package lsmeans. J. Stat. Softw. 2016, 69, 1–33. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, J.; Wishart, D.S. MSEA: A web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010, 38, W71–W77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Jewison, T.; Su, Y.; Disfany, F.M.; Liang, Y.; Knox, C.; Maciejewski, A.; Poelzer, J.; Huynh, J.; Zhou, Y.; Arndt, D.; et al. SMPDB 2.0: Big improvements to the Small Molecule Pathway Database. Nucleic Acids Res. 2014, 42, D478–D484. [Google Scholar] [CrossRef] [Green Version]
- Pedroso, A.P.; Souza, A.P.; Dornellas, A.P.S.; Oyama, L.M.; Nascimento, C.M.O.; Santos, G.M.S.; Rosa, J.C.; Bertolla, R.P.; Klawitter, J.; Christians, U.; et al. Intrauterine Growth Restriction Programs the Hypothalamus of Adult Male Rats: Integrated Analysis of Proteomic and Metabolomic Data. J. Proteome Res. 2017, 16, 1515–1525. [Google Scholar] [CrossRef]
- Pedroso, A.P.; Dornellas, A.P.S.; de Souza, A.P.; Pagotto, J.F.; Oyama, L.M.; Nascimento, C.M.O.; Klawitter, J.; Christians, U.; Tashima, A.K.; Ribeiro, E.B. A proteomics-metabolomics approach indicates changes in hypothalamic glutamate-GABA metabolism of adult female rats submitted to intrauterine growth restriction. Eur. J. Nutr. 2019, 58, 3059–3068. [Google Scholar] [CrossRef] [Green Version]
- Palouzier-Paulignan, B.; Lacroix, M.-C.; Aimé, P.; Baly, C.; Caillol, M.; Congar, P.; Julliard, A.K.; Tucker, K.; Fadool, D.A. Olfaction under metabolic influences. Chem. Senses 2012, 37, 769–797. [Google Scholar] [CrossRef]
- Li, L.O.; Hu, Y.-F.; Wang, L.; Mitchell, M.; Berger, A.; Coleman, R.A. Early hepatic insulin resistance in mice: A metabolomics analysis. Mol. Endocrinol. Baltim. Md 2010, 24, 657–666. [Google Scholar] [CrossRef] [Green Version]
- Yamanouchi, T.; Akanuma, H.; Takaku, F.; Akanuma, Y. Marked depletion of plasma 1,5-anhydroglucitol, a major polyol, in streptozocin-induced diabetes in rats and the effect of insulin treatment. Diabetes 1986, 35, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.J.; Park, C.-Y. 1,5-Anhydroglucitol in diabetes mellitus. Endocrine 2013, 43, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. Biochemistry; W.H. Freeman: Basingstoke, UK, 2012; ISBN 978-1-4292-7635-1. [Google Scholar]
- Roy, M.; Hennebelle, M.; St-Pierre, V.; Courchesne-Loyer, A.; Fortier, M.; Bouzier-Sore, A.-K.; Gallis, J.-L.; Beauvieux, M.-C.; Cunnane, S.C. Long-term calorie restriction has minimal impact on brain metabolite and fatty acid profiles in aged rats on a Western-style diet. Neurochem. Int. 2013, 63, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, L.; Geller, S.; Fioramonti, X.; Hébert, A.; Repond, C.; Leloup, C.; Pellerin, L. Evidence for hypothalamic ketone body sensing: Impact on food intake and peripheral metabolic responses in mice. Am. J. Physiol. Endocrinol. Metab. 2016, 310, E103–E115. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Kim, D.S.; Daily, J.W. Central infusion of ketone bodies modulates body weight and hepatic insulin sensitivity by modifying hypothalamic leptin and insulin signaling pathways in type 2 diabetic rats. Brain Res. 2011, 1401, 95–103. [Google Scholar] [CrossRef]
- Juge, N.; Gray, J.A.; Omote, H.; Miyaji, T.; Inoue, T.; Hara, C.; Uneyama, H.; Edwards, R.H.; Nicoll, R.A.; Moriyama, Y. Metabolic control of vesicular glutamate transport and release. Neuron 2010, 68, 99–112. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.; Berg, J.; Yellen, G. Ketogenic diet metabolites reduce firing in central neurons by opening K(ATP) channels. J. Neurosci. Off. J. Soc. Neurosci. 2007, 27, 3618–3625. [Google Scholar] [CrossRef] [Green Version]
- Lacroix, M.-C.; Caillol, M.; Durieux, D.; Monnerie, R.; Grebert, D.; Pellerin, L.; Repond, C.; Tolle, V.; Zizzari, P.; Baly, C. Long-Lasting Metabolic Imbalance Related to Obesity Alters Olfactory Tissue Homeostasis and Impairs Olfactory-Driven Behaviors. Chem. Senses 2015, 40, 537–556. [Google Scholar] [CrossRef]
- Le Foll, C.; Dunn-Meynell, A.A.; Miziorko, H.M.; Levin, B.E. Role of VMH ketone bodies in adjusting caloric intake to increased dietary fat content in DIO and DR rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015, 308, R872–R878. [Google Scholar] [CrossRef] [Green Version]
- Rojas-Morales, P.; Tapia, E.; Pedraza-Chaverri, J. β-Hydroxybutyrate: A signaling metabolite in starvation response? Cell. Signal. 2016, 28, 917–923. [Google Scholar] [CrossRef]
- Wells, A.; Barrington, W.T.; Dearth, S.; May, A.; Threadgill, D.W.; Campagna, S.R.; Voy, B.H. Tissue Level Diet and Sex-by-Diet Interactions Reveal Unique Metabolite and Clustering Profiles Using Untargeted Liquid Chromatography-Mass Spectrometry on Adipose, Skeletal Muscle, and Liver Tissue in C57BL6/J Mice. J. Proteome Res. 2018, 17, 1077–1090. [Google Scholar] [CrossRef] [PubMed]
- Soltis, A.R.; Kennedy, N.J.; Xin, X.; Zhou, F.; Ficarro, S.B.; Yap, Y.S.; Matthews, B.J.; Lauffenburger, D.A.; White, F.M.; Marto, J.A.; et al. Hepatic Dysfunction Caused by Consumption of a High-Fat Diet. Cell Rep. 2017, 21, 3317–3328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.; Dzierlenga, A.L.; Lu, Z.; Billheimer, D.D.; Torabzadeh, E.; Lake, A.D.; Li, H.; Novak, P.; Shipkova, P.; Aranibar, N.; et al. Metabolomic profiling distinction of human nonalcoholic fatty liver disease progression from a common rat model. Obes. Silver Spring Md 2017, 25, 1069–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allam-Ndoul, B.; Guénard, F.; Garneau, V.; Cormier, H.; Barbier, O.; Pérusse, L.; Vohl, M.-C. Association between Metabolite Profiles, Metabolic Syndrome and Obesity Status. Nutrients 2016, 8, 324. [Google Scholar] [CrossRef] [Green Version]
- Goffredo, M.; Santoro, N.; Tricò, D.; Giannini, C.; D’Adamo, E.; Zhao, H.; Peng, G.; Yu, X.; Lam, T.T.; Pierpont, B.; et al. A Branched-Chain Amino Acid-Related Metabolic Signature Characterizes Obese Adolescents with Non-Alcoholic Fatty Liver Disease. Nutrients 2017, 9, 642. [Google Scholar] [CrossRef] [Green Version]
- Hadjihambi, A.; Arias, N.; Sheikh, M.; Jalan, R. Hepatic encephalopathy: A critical current review. Hepatol. Int. 2018, 12, 135–147. [Google Scholar] [CrossRef] [Green Version]
- Hassan, A.M.; Mancano, G.; Kashofer, K.; Fröhlich, E.E.; Matak, A.; Mayerhofer, R.; Reichmann, F.; Olivares, M.; Neyrinck, A.M.; Delzenne, N.M.; et al. High-fat diet induces depression-like behaviour in mice associated with changes in microbiome, neuropeptide Y, and brain metabolome. Nutr. Neurosci. 2019, 22, 877–893. [Google Scholar] [CrossRef] [Green Version]
- Crowther, L.M.; Mathis, D.; Poms, M.; Plecko, B. New insights into human lysine degradation pathways with relevance to pyridoxine-dependent epilepsy due to antiquitin deficiency. J. Inherit. Metab. Dis. 2019, 42, 620–628. [Google Scholar] [CrossRef] [Green Version]
- Papes, F.; Surpili, M.J.; Langone, F.; Trigo, J.R.; Arruda, P. The essential amino acid lysine acts as precursor of glutamate in the mammalian central nervous system. FEBS Lett. 2001, 488, 34–38. [Google Scholar] [CrossRef] [Green Version]
- Weiss, N.; Barbier Saint Hilaire, P.; Colsch, B.; Isnard, F.; Attala, S.; Schaefer, A.; Amador, M.D.M.; Rudler, M.; Lamari, F.; Sedel, F.; et al. Cerebrospinal fluid metabolomics highlights dysregulation of energy metabolism in overt hepatic encephalopathy. J. Hepatol. 2016, 65, 1120–1130. [Google Scholar] [CrossRef]
- Julliard, A.-K.; Al Koborssy, D.; Fadool, D.A.; Palouzier-Paulignan, B. Nutrient Sensing: Another Chemosensitivity of the Olfactory System. Front. Physiol. 2017, 8, 468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazutkaite, G.; Soldà, A.; Lossow, K.; Meyerhof, W.; Dale, N. Amino acid sensing in hypothalamic tanycytes via umami taste receptors. Mol. Metab. 2017, 6, 1480–1492. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Docosahexaenoic Acid. Ann. Nutr. Metab. 2016, 69 (Suppl. 1), 7–21. [Google Scholar] [CrossRef] [PubMed]
- Domenichiello, A.F.; Kitson, A.P.; Bazinet, R.P. Is docosahexaenoic acid synthesis from α-linolenic acid sufficient to supply the adult brain? Prog. Lipid Res. 2015, 59, 54–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fekete, K.; Györei, E.; Lohner, S.; Verduci, E.; Agostoni, C.; Decsi, T. Long-chain polyunsaturated fatty acid status in obesity: A systematic review and meta-analysis. Obes. Rev. Off. J. Int. Assoc. Study Obes. 2015, 16, 488–497. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-López, M.T.; Vázquez, M.; Lomazzo, E.; Hofmann, C.; Blanco, R.N.; Alén, F.; Antón, M.; Decara, J.; Arco, R.; Orio, L.; et al. A moderate diet restriction during pregnancy alters the levels of endocannabinoids and endocannabinoid-related lipids in the hypothalamus, hippocampus and olfactory bulb of rat offspring in a sex-specific manner. PLoS ONE 2017, 12, e0174307. [Google Scholar] [CrossRef]
- Hichami, A.; Datiche, F.; Ullah, S.; Liénard, F.; Chardigny, J.-M.; Cattarelli, M.; Khan, N.A. Olfactory discrimination ability and brain expression of c-fos, Gir and Glut1 mRNA are altered in n-3 fatty acid-depleted rats. Behav. Brain Res. 2007, 184, 1–10. [Google Scholar] [CrossRef]
- Alexandre-Gouabau, M.-C.; Courant, F.; Le Gall, G.; Moyon, T.; Darmaun, D.; Parnet, P.; Coupé, B.; Antignac, J.-P. Offspring metabolomic response to maternal protein restriction in a rat model of intrauterine growth restriction (IUGR). J. Proteome Res. 2011, 10, 3292–3302. [Google Scholar] [CrossRef]
- Paul, H.A.; Collins, K.H.; Bomhof, M.R.; Vogel, H.J.; Reimer, R.A. Potential Impact of Metabolic and Gut Microbial Response to Pregnancy and Lactation in Lean and Diet-Induced Obese Rats on Offspring Obesity Risk. Mol. Nutr. Food Res. 2018, 62. [Google Scholar] [CrossRef]
- Rauschert, S.; Kirchberg, F.F.; Marchioro, L.; Koletzko, B.; Hellmuth, C.; Uhl, O. Early Programming of Obesity Throughout the Life Course: A Metabolomics Perspective. Ann. Nutr. Metab. 2017, 70, 201–209. [Google Scholar] [CrossRef]
- Pereira, T.J.; Fonseca, M.A.; Campbell, K.E.; Moyce, B.L.; Cole, L.K.; Hatch, G.M.; Doucette, C.A.; Klein, J.; Aliani, M.; Dolinsky, V.W. Maternal obesity characterized by gestational diabetes increases the susceptibility of rat offspring to hepatic steatosis via a disrupted liver metabolome. J. Physiol. 2015, 593, 3181–3197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wesolowski, S.R.; Mulligan, C.M.; Janssen, R.C.; Baker, P.R.; Bergman, B.C.; D’Alessandro, A.; Nemkov, T.; Maclean, K.N.; Jiang, H.; Dean, T.A.; et al. Switching obese mothers to a healthy diet improves fetal hypoxemia, hepatic metabolites, and lipotoxicity in non-human primates. Mol. Metab. 2018, 18, 25–41. [Google Scholar] [CrossRef] [PubMed]
- Kubomura, D.; Matahira, Y.; Masui, A.; Matsuda, H. Intestinal absorption and blood clearance of L-histidine-related compounds after ingestion of anserine in humans and comparison to anserine-containing diets. J. Agric. Food Chem. 2009, 57, 1781–1785. [Google Scholar] [CrossRef] [PubMed]
- McManus, R. Enzymatic Synthesis of Anserine in Skeletal Muscle by N-Methylation of Carnosine. J. Biol. Chem. 1962, 237, 1207–1211. [Google Scholar]
- Chen, P.-J.; Tseng, J.-K.; Lin, Y.-L.; Wu, Y.-H.S.; Hsiao, Y.-T.; Chen, J.-W.; Chen, Y.-C. Protective Effects of Functional Chicken Liver Hydrolysates against Liver Fibrogenesis: Antioxidation, Anti-inflammation, and Antifibrosis. J. Agric. Food Chem. 2017, 65, 4961–4969. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.-H.; Wang, S.-Y.; Lin, Y.-T.; Chen, Y.-C. Antioxidant activities of chicken liver hydrolysates by pepsin treatment. Int. J. Food Sci. Technol. 2014, 49, 1654–1662. [Google Scholar] [CrossRef]
- Mong, M.; Chao, C.; Yin, M. Histidine and carnosine alleviated hepatic steatosis in mice consumed high saturated fat diet. Eur. J. Pharmacol. 2011, 653, 82–88. [Google Scholar] [CrossRef]
- Yang, K.-T.; Lin, C.; Liu, C.-W.; Chen, Y.-C. Effects of chicken-liver hydrolysates on lipid metabolism in a high-fat diet. Food Chem. 2014, 160, 148–156. [Google Scholar] [CrossRef]





| Decreased under HFD | Increased under HFD | |
|---|---|---|
| Liver (LI) | 1,5-Anhydro-d-sorbitol; l-Saccharopine; Indoxyl-sulfate; Imidazolelactic-acid; Methylhippurate; 3-Methylhistidine; Methionine; 2-Deoxyribose-5-phosphate; UMP; Inosine-5-monophosphate; GMP; 4-Guanidinobutyric-acid; 3-AMP/5-AMP; 4-Hydroxy-3-methoxyphenylglycol-sulfate; 3-Methylcrotonyl-glycine/N-Tiglylglycine; Guanine; l-Alanyl-l-proline; d-Glucosamine-6-phosphate; Glycerol-3-phosphate; Hydroxyphenylpyruvic-acid; Cytidine; ortho-Methylhippuric-acid/meta-Methylhippuric-acid/para-Methylhippuric-acid/Phenylacetylglycine; d-Fructose-1-phosphate/d-Fructose-6-phosphate/d-Mannose-6-phosphate/Galactose-1-phosphate/Glucose-1-phosphate/Glucose-6-PO4/Mannose-1-phosphate; Caprylolyglycine; UDP; Nervonic-acid; N-Acetyl-l-methionine; N-Isobutyrylglycine; 2-Oxobutyric-acid; N-Glycolylneuraminic-acid; 5-AMP/dGMP; Phosphonoacetic-acid; Guanosine-5-diphospho-d-mannose/Guanosine-5-diphosphoglucose; dGMP; Serine; Orotic-acid; Hexanoyl-glycine/N-Acetyl-d-allo-isoleucine/N-Acetyl-l-leucine; N-Acetyl-d-penicillamine/N-Acetyl-l-Methionine; Hexanoyl-glycine; Phosphoserine; Deoxyinosine; Uridine; Sebacic-acid; Guanosine-5-diphospho-l-fucose; 5-Aminolevulinic-acid/cis-4-hydroxy-d-proline/trans-3-Hydroxy-l-proline/trans-4-hydroxy-l-proline; Purine; Myristic-acid; N-Acetylneuraminic-acid; d-Pyroglutamic-acid; Muramic-acid; Leu-Pro; Pyrrole-2-carboxylic-acid; D-Arabinose | 3-Hydroxybutyric-acid; N-N-Dimethylglycine; Betaine; NAD; Taurine; 13-S-Hydroxyoctadeca-9Z-11E-dienoic-acid; Argininosuccinic-acid; 1-Methyladenosine; S-Adenosyl-homocysteine; Cyclic-ADP-ribose; dl-alpha-Hydroxystearic-acid; gamma-Linolenic-acid; β-Alanine; d-Threitol; Asparagine; 3-Ureidopropionic-acid; Quinolinic-acid; l-Glutamic-acid; N-Methyl-d-aspartic-acid; Stachydrine; l-Cysteinesulfinic-acid; l-Kynurenine; Propionylcarnitine; ADP; 2-O-Methylinosine; Riboflavin; Pyridoxamine-5-phosphate; Sphinganine; 3-Hydroxy-2-methyl-butanoic-acid/3-Hydroxypentanoic-acid; Perillic-acid; l-Cysteic-acid; d-Sphingosine; 2-Aminopyridine-3-carboxylic-acid; Phosphoenolpyruvic-acid; 2-O-Methylguanosine; Fumaric-acid/Maleic-acid; 3-Hydroxypicolinic-acid; Uracil; Xanthosine; Diglycolic-acid/Malic-acid; d-Mannitol-1-phosphate; Prostaglandin-A1; Carnitine; Prostaglandin-E1; Indolelactic-acid; 5-Aminoimidazole-4-carboxamide-1b-d-ribofuranoside; Nicotinic-acid; 2-Hydroxyhexadecanoic-acid; Xanthine; N6-N6-N6-Trimethyl-l-lysine; Arachidic-acid; Cytidine-5-diphosphocholine; 4-Pyridoxic-acid; d-Glyceric-acid; Aldosterone |
| Hypothalamus (HYP) | 1,5-Anhydro-d-sorbitol; l-Saccharopine; 3-Methylhistidine | 3-Hydroxybutyric-acid; Argininosuccinic-acid; Pantothenic-acid; Tyrosine; dl-Tryptophan; Phenylalanine; Arginine; Tartaric-acid; Valine; Asparagine; Gly-Pro/Pro-Gly; 3-Aminosalicylic-acid; Leu-Pro |
| Whole olfactory bulb (WOB) | 1,5-Anhydro-d-sorbitol; l-Saccharopine; cis-5,8,11,14,17-Eicosapentaenoic-acid; cis-8,11,14-Eicosatrienoic-acid; Lysine; trans-4-hydroxy-l-proline; l-Homoserine/Threonine | 3-Hydroxybutyric-acid; gamma-Linolenic-acid; Pantothenic-acid; 3-Aminosalicylic-acid |
| Pathways | Total Compounds | Hits | FDR | Impact | Metabolites |
|---|---|---|---|---|---|
| D-Glutamine and D-glutamate metabolism | 5 | 1 | 0.00002 | 100% | L-Glutamic acid ↑ |
| Ubiquinone and other terpenoid-quinone biosynthesis | 3 | 1 | 0.00644 | 100% | 4-Hydroxyphenylpyruvic acid ↓ |
| Taurine and hypotaurine metabolism | 8 | 3 | 0.00002 | 71% | Cysteic acid ↑; 3-Sulfinoalanine ↑; Taurine ↑ |
| Beta-alanine metabolism | 17 | 3 | 0.00001 | 67% | Beta-alanine ↑; Ureidopropionic acid ↑; Uracil ↑ |
| Methane metabolism | 9 | 1 | 0.02049 | 40% | L-Serine ↓ |
| Glycine, serine and threonine metabolism | 31 | 5 | 0.00001 | 36% | Dimethylglycine ↑; Phosphoserine ↓ |
| Purine metabolism | 68 | 8 | 0.00001 | 28% | Xanthine ↑; AICAR ↑; Inosinic acid ↓; Deoxyinosine ↓; Xanthosine ↑; Guanosine monophosphate ↓; Guanine ↓; 2′-Deoxyguanosine 5′-monophosphate ↓ |
| Pyrimidine metabolism | 41 | 8 | 0.00001 | 28% | Uridine 5′-diphosphate ↓; Uridine 5′-monophosphate ↓; Uridine ↑; Ureidopropionic acid ↑; Cytidine ↑; Orotic acid ↑; Uracil ↑; Beta-Alanine ↑ |
| Alanine, aspartate and glutamate metabolism | 24 | 4 | 0.00001 | 28% | Argininosuccinic acid ↑; L-Glutamic acid ↑; L-Asparagine ↓; Glucosamine 6-phosphate ↓ |
| Nicotinate and nicotinamide metabolism | 13 | 3 | 0.00001 | 21% | Quinolinic acid ↑; NAD ↑; Nicotinic acid ↑ |
| Cysteine and methionine metabolism | 27 | 6 | 0.00001 | 20% | L-Serine ↓; L-Methionine ↑; S-Adenosylhomocysteine ↑; Cysteic acid ↑; 3-Sulfinoalanine ↑; 2-Ketobutyric acid ↓ |
| Sphingolipid metabolism | 21 | 3 | 0.00005 | 20% | Sphinganine ↑; L-Serine ↓; Sphingosine ↑ |
| Glycerolipid metabolism | 18 | 2 | 0.00019 | 13% | Glycerol 3-phosphate ↓; Glyceric acid ↑ |
| Glycerophospholipid metabolism | 30 | 2 | 0.00012 | 13% | Citicoline ↑; Glycerol 3-phosphate ↓ |
| Aminoacyl-tRNA biosynthesis | 69 | 4 | 0.00038 | 13% | L-Asparagine ↓; L-Serine ↓; L-Methionine ↑; L-Glutamic acid ↑ |
| Arginine and proline metabolism | 44 | 3 | 0.00001 | 12% | Argininosuccinic acid ↑; L-Glutamic acid ↑; 4-Guanidinobutanoic acid ↓ |
| Tryptophan metabolism | 40 | 1 | 0.00056 | 11% | L-Kynurenine ↓ |
| Glycolysis or gluconeogenesis | 26 | 1 | 0.00032 | 10% | Phosphoenolpyruvic acid ↑ |
| Amino sugar and nucleotide sugar metabolism | 37 | 3 | 0.00020 | 8% | Glucosamine 6-phosphate ↓; GDP-L-fucose ↓; N-Glycolylneuraminic acid ↓ |
| Tyrosine metabolism | 44 | 1 | 0.00645 | 7% | 4-Hydroxyphenylpyruvic acid ↓ |
| Pentose phosphate pathway | 19 | 1 | 0.00002 | 7% | Deoxyribose 5-phosphate ↓ |
| Glyoxylate and dicarboxylate metabolism | 18 | 1 | 0.00032 | 6% | Glyceric acid ↑ |
| Glutathione metabolism | 26 | 1 | 0.00002 | 6% | L-Glutamic acid ↑ |
| Vitamin B6 metabolism | 9 | 2 | 0.00002 | 5% | Pyridoxamine 5′-phosphate ↑; 4-Pyridoxic acid ↑ |
| Pantothenate and CoA biosynthesis | 15 | 3 | 0.00001 | 4% | Ureidopropionic acid ↑; Beta-Alanine ↑; Uracil ↑ |
| Primary bile acid biosynthesis | 46 | 1 | 0.00002 | 3% | Taurine ↑ |
| Lysine degradation | 23 | 2 | 0.00001 | 1% | N6,N6,N6-Trimethyl-L-lysine ↑; Saccharopine ↓ |
| Histidine metabolism | 15 | 2 | 0.00001 | 0% | L-Glutamic acid ↑; 1-Methylhistidine ↓ |
| Biosynthesis of unsaturated fatty acids | 42 | 3 | 0.00001 | 0% | Nervonic acid ↓; Arachidic acid ↑; Gamma-Linolenic acid ↑ |
| Butanoate metabolism | 22 | 1 | 0.00002 | 0% | L-Glutamic acid ↑ |
| Porphyrin and chlorophyll metabolism | 27 | 1 | 0.00002 | 0% | L-Glutamic acid ↑ |
| Nitrogen metabolism | 9 | 1 | 0.00002 | 0% | L-Glutamic acid ↑ |
| Propanoate metabolism | 20 | 2 | 0.00003 | 0% | Beta-Alanine ↑; 2-Ketobutyric acid ↓ |
| Linoleic acid metabolism | 6 | 1 | 0.00003 | 0% | 13S-hydroxyoctadecadienoic acid ↑ |
| Limonene and pinene degradation | 8 | 1 | 0.00011 | 0% | Perillic acid ↑ |
| Riboflavin metabolism | 11 | 1 | 0.00012 | 0% | Riboflavin ↑ |
| Citrate cycle (TCA cycle) | 20 | 1 | 0.00032 | 0% | Phosphoenolpyruvic acid ↑ |
| Pyruvate metabolism | 23 | 1 | 0.00032 | 0% | Phosphoenolpyruvic acid ↑ |
| Lysine biosynthesis | 4 | 1 | 0.00466 | 0% | Saccharopine ↓ |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 4 | 1 | 0.00645 | 0% | 4-Hydroxyphenylpyruvic acid ↓ |
| Cyanoamino acid metabolism | 6 | 1 | 0.02049 | 0% | L-Serine ↓ |
| Pathways | Total Compounds | Hits | FDR | Impact | Metabolites |
|---|---|---|---|---|---|
| Hypothalamus (KEGG and SMPDB databases retrieved the same results) | |||||
| Aspartate metabolism | 34 | 3 | 0.00818 | 35% | L-Asparagine ↑, Argininosuccinic acid ↑, L-Arginine ↑ |
| Arginine and proline metabolism | 48 | 2 | 0.00586 | 24% | Argininosuccinic acid ↑, L-Arginine ↑ |
| Urea cycle | 23 | 2 | 0.00586 | 22% | Argininosuccinic acid ↑, L-Arginine ↑ |
| Phenylalanine and tyrosine metabolism | 25 | 2 | 0.01921 | 12% | L-Phenylalanine ↑, L-Tyrosine ↑ |
| Pantothenate and CoA biosynthesis | 19 | 1 | 0.00586 | 7% | Pantothenic acid ↑ |
| Ammonia recycling | 25 | 1 | 0.03670 | 3% | L-Asparagine ↑ |
| Lysine degradation | 20 | 1 | 0.01921 | 3% | Saccharopine ↓ |
| Beta-alanine metabolism | 26 | 2 | 0.00586 | 0% | 3-Methylhistidine ↑, Pantothenic acid ↑ |
| Valine, leucine and isoleucine degradation | 51 | 1 | 0.02030 | 0% | L-Valine ↑ |
| Catecholamine biosynthesis | 14 | 1 | 0.03065 | 0% | L-Tyrosine ↑ |
| Tyrosine metabolism | 55 | 1 | 0.03065 | 0% | L-Tyrosine ↑ |
| Whole olfactory bulb (KEGG database) | |||||
| Arginine and proline metabolism | 44 | 1 | 0.00233 | 4% | Hydroxyproline ↓ |
| Pantothenate and CoA biosynthesis | 15 | 1 | 0.00430 | 2% | Pantothenic acid ↑ |
| Lysine degradation | 23 | 2 | 0.00015 | 1% | L-Lysine ↓; Saccharopine ↓ |
| Biosynthesis of unsaturated fatty acids | 42 | 3 | 0.00015 | 0% | 8,11,14-Eicosatrienoic acid ↓; Gamma-linolenic acid ↑; Eicosapentaenoic acid ↓ |
| Lysine biosynthesis | 4 | 2 | 0.00015 | 0% | L-Lysine ↓; Saccharopine ↓ |
| Biotin metabolism | 5 | 1 | 0.00430 | 0% | L-Lysine ↓ |
| Aminoacyl-tRNA biosynthesis | 69 | 1 | 0.00430 | 0% | L-Lysine ↓ |
| Whole olfactory bulb (SMPDB database) | |||||
| Alpha linolenic acid and linoleic acid metabolism | 17 | 3 | 0.00032 | 26% | Eicosapentaenoic acid ↓; 8,11,14-Eicosatrienoic acid ↓; Gamma-Linolenic acid ↑ |
| Lysine degradation | 20 | 2 | 0.18397 | 3% | L-Lysine ↓; Saccharopine ↓ |
| Biotin metabolism | 7 | 1 | 0.72255 | 0% | L-Lysine ↓ |
| Carnitine synthesis | 16 | 1 | 0.72255 | 0% | L-Lysine ↓ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Safi-Stibler, S.; Thévenot, E.A.; Jouneau, L.; Jouin, M.; Seyer, A.; Jammes, H.; Rousseau-Ralliard, D.; Baly, C.; Gabory, A. Differential Effects of Post-Weaning Diet and Maternal Obesity on Mouse Liver and Brain Metabolomes. Nutrients 2020, 12, 1572. https://doi.org/10.3390/nu12061572
Safi-Stibler S, Thévenot EA, Jouneau L, Jouin M, Seyer A, Jammes H, Rousseau-Ralliard D, Baly C, Gabory A. Differential Effects of Post-Weaning Diet and Maternal Obesity on Mouse Liver and Brain Metabolomes. Nutrients. 2020; 12(6):1572. https://doi.org/10.3390/nu12061572
Chicago/Turabian StyleSafi-Stibler, Sofiane, Etienne A. Thévenot, Luc Jouneau, Mélanie Jouin, Alexandre Seyer, Hélène Jammes, Delphine Rousseau-Ralliard, Christine Baly, and Anne Gabory. 2020. "Differential Effects of Post-Weaning Diet and Maternal Obesity on Mouse Liver and Brain Metabolomes" Nutrients 12, no. 6: 1572. https://doi.org/10.3390/nu12061572