Urolithin Metabotypes can Anticipate the Different Restoration of the Gut Microbiota and Anthropometric Profiles during the First Year Postpartum
Abstract
:1. Introduction
2. Materials and Methods
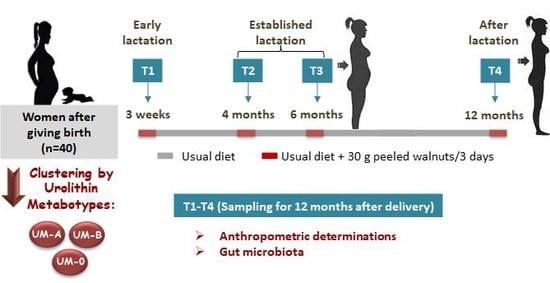
2.1. Study Design and Diet
2.2. Anthropometric Determinations
2.3. Sampling Procedure and Determinations
2.4. Statistical Analysis
3. Results
3.1. Distribution of Microbiome Enterotypes and Urolithin Metabotypes (UMs) of the Mothers Over One Year After Delivery
3.2. Changes of Anthropometric Values over One Year after Giving Birth
3.3. Alpha-Diversity and Composition of the Maternal Gut Microbiota for One Year after Delivery
3.4. The Functionality of the Maternal Gut Microbiota During the First Year after Delivery
3.5. Associations of Gut Microbial and Anthropometric Changes of the Mothers Over One Year after Delivery
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gregor, M.F.; Hotamisligil, G.S. Inflammatory Mechanisms in Obesity. Annu. Rev. Immunol. 2011, 29, 415–445. [Google Scholar] [CrossRef] [Green Version]
- Kalliomäki, M.; Collado, M.C.; Salminen, S.; Isolauri, E. Early differences in fecal microbiota composition in children may predict overweight. Am. J. Clin. Nutr. 2008, 87, 534–538. [Google Scholar] [CrossRef]
- Koren, O.; Goodrich, J.K.; Cullender, T.C.; Spor, A.A.; Laitinen, K.; Backhed, H.K.; Gonzalez, A.; Werner, J.J.; Angenent, L.T.; Knight, R.; et al. During Pregnancy. Cell 2013, 150, 470–480. [Google Scholar] [CrossRef]
- Dalrymple, K.V.; Flynn, A.C.; Relph, S.A.; O’Keeffe, M.; Poston, L. Lifestyle Interventions in Overweight and Obese Pregnant or Postpartum Women for Postpartum Weight Management: A Systematic Review of the Literature. Nutrients 2018, 10, 1704. [Google Scholar] [CrossRef]
- Endres, L.K.; Straub, H.; McKinney, C.; Plunkett, B.; Minkovitz, C.S.; Schetter, C.D.; Ramey, S.; Wang, C.; Hobel, C.; Raju, T.; et al. Postpartum weight retention risk factors and relationship to obesity at 1 year. Obstet. Gynecol. 2015, 125, 144–152. [Google Scholar] [CrossRef]
- Benson, A.K.; Kelly, S.A.; Legge, R.; Ma, F.; Low, S.J.; Kim, J.; Zhang, M.; Oh, P.L.; Nehrenberg, D.; Hua, K.; et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18933–18938. [Google Scholar] [CrossRef] [Green Version]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.-Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef]
- Zhou, Y.; Mihindukulasuriya, K.A.; Gao, H.; La Rosa, P.S.; Wylie, K.M.; Martin, J.C.; Kota, K.; Shannon, W.D.; Mitreva, M.; Sodergren, E.; et al. Exploration of bacterial community classes in major human habitats. Genome Biol. 2014, 15, R66. [Google Scholar] [CrossRef]
- Costea, P.I.; Hildebrand, F.; Arumugam, M.; Bäckhed, F.; Blaser, M.J.; Bushman, F.D.; Bork, P. Enterotypes in the landscape of gut microbial community composition. Nat. Microbiol. 2018, 3, 8–16. [Google Scholar] [CrossRef]
- Tomas-Barberan, F.A.; Selma, M.V.; Espín, J.C. Polyphenols’ Gut Microbiota Metabolites: Bioactives or Biomarkers? J. Agric. Food Chem. 2018, 66. [Google Scholar] [CrossRef]
- Espín, J.C.; González-Sarrías, A.; Tomás-Barberán, F.A. The gut microbiota: A key factor in the therapeutic effects of (poly)phenols. Biochem. Pharmacol. 2017, 139, 82–93. [Google Scholar] [CrossRef]
- Tomás-Barberán, F.A.; García-Villalba, R.; González-Sarrías, A.; Selma, M.V.; Espín, J.C. Ellagic acid metabolism by human gut microbiota: Consistent observation of three urolithin phenotypes in intervention trials, independent of food source, age, and health status. J. Agric. Food Chem. 2014, 62, 6535–6538. [Google Scholar] [CrossRef]
- Tomás-Barberán, F.A.; González-Sarrías, A.; García-Villalba, R.; Núñez-Sánchez, M.A.; Selma, M.V.; García-Conesa, M.T.; Espín, J.C. Urolithins, the rescue of “old” metabolites to understand a “new” concept: Metabotypes as a nexus among phenolic metabolism, microbiota dysbiosis, and host health status. Mol. Nutr. Food Res. 2017, 61, 1500901. [Google Scholar] [CrossRef]
- Selma, M.V.; Beltrán, D.; García-Villalba, R.; Espín, J.C.; Tomás-Barberán, F.A. Description of urolithin production capacity from ellagic acid of two human intestinal Gordonibacter species. Food Func. 2014, 5, 1779–1784. [Google Scholar] [CrossRef] [Green Version]
- Selma, M.V.; Tomas-Barberan, F.A.; Beltran, D.; Garcia-Villalba, R.; Espin, J.C. Gordonibacter urolithinfaciens sp. nov., a urolithin-producing bacterium isolated from the human gut. Int. J. Syst. Evol. Microbiol. 2014, 64, 2346–2352. [Google Scholar] [CrossRef]
- Selma, M.V.; Beltrán, D.; Luna, M.C.; Romo-Vaquero, M.; García-Villalba, R.; Mira, A.; Espín, J.C.; Tomás-Barberán, F.A. Isolation of human intestinal bacteria capable of producing the bioactive metabolite isourolithin a from ellagic acid. Front. Microbiol. 2017, 8, 1–8. [Google Scholar] [CrossRef]
- Beltrán, D.; Romo-Vaquero, M.; Espín, J.C.; Tomás-Barberán, F.A.; Selma, M.V. Ellagibacter isourolithinifaciens gen. Nov., sp. Nov., a new member of the family Eggerthellaceae, isolated from human gut. Int. J. Syst. Evol. Microbiol. 2018, 68, 1707–1712. [Google Scholar] [CrossRef]
- Romo-Vaquero, M.; Cortés-Martín, A.; Loria-Kohen, V.; Ramírez-de-Molina, A.; García-Mantrana, I.; Collado, M.C.; Espín, J.C.; Selma, M.V. Deciphering the Human Gut Microbiome of Urolithin Metabotypes: Association with Enterotypes and Potential Cardiometabolic Health Implications. Mol. Nutr. Food Res. 2019, 63, 1–11. [Google Scholar] [CrossRef]
- González-Sarrías, A.; Romo-Vaquero, M.; García-Villalba, R.; Cortés-Martín, A.; Selma, M.V.; Espín, J.C. The Endotoxemia Marker Lipopolysaccharide-Binding Protein is Reduced in Overweight-Obese Subjects Consuming Pomegranate Extract by Modulating the Gut Microbiota: A Randomized Clinical Trial. Mol. Nutr. Food Res. 2018, 4, 1800160. [Google Scholar] [CrossRef]
- Selma, M.V.; González-Sarrías, A.; Salas-Salvadó, J.; Andrés-Lacueva, C.; Alasalvar, C.; Örem, A.; Tomás-Barberán, F.A.; Espín, J.C. The gut microbiota metabolism of pomegranate or walnut ellagitannins yields two urolithin-metabotypes that correlate with cardiometabolic risk biomarkers: Comparison between normoweight, overweight-obesity and metabolic syndrome. Clin. Nutr. 2018, 37, 897–905. [Google Scholar] [CrossRef]
- Selma, M.V.; Romo-Vaquero, M.; García-Villalba, R.; González-Sarrías, A.; Tomás-Barberán, F.A.; Espín, J.C. The human gut microbial ecology associated with overweight and obesity determines ellagic acid metabolism. Food Func. 2016, 1769–1774. [Google Scholar] [CrossRef]
- Cortés-Martín, A.; García-Villalba, R.; González-Sarrías, A.; Romo-Vaquero, M.; Loria-Kohen, V.; Ramírez-De-Molina, A.; Tomás-Barberán, F.A.; Selma, M.V.; Espín, J.C. The gut microbiota urolithin metabotypes revisited: The human metabolism of ellagic acid is mainly determined by aging. Food Funct. 2018, 9, 4100–4106. [Google Scholar] [CrossRef]
- Martínez-González, M.Á.; Corella, D.; Salas-Salvadó, J.; Ros, E.; Covas, M.I.; Fiol, M.; Wärnberg, J.; Arós, F.; Ruíz-Gutiérrez, V.; Lamuela-Raventós, R.M.; et al. Cohort Profile: Design and methods of the PREDIMED study. Int. J. Epidemiol. 2012, 41, 377–385. [Google Scholar] [CrossRef]
- WHO. Obesity: Preventing and Managing the Global Epidemic. Report of a WHO Consultation; World Health Organization Technical Report Series 894; WHO: Geneva, Switzerland, 2000; pp. 1–253. [Google Scholar]
- García-Villalba, R.; Espín, J.C.; Tomás-Barberán, F.A. Chromatographic and spectroscopic characterization of urolithins for their determination in biological samples after the intake of foods containing ellagitannins and ellagic acid. J. Chromatogr. A 2015, 1428, 162–175. [Google Scholar] [CrossRef]
- Byerley, L.O.; Samuelson, D.; Blanchard, E.; Luo, M.; Lorenzen, B.N.; Banks, S.; Ponder, M.A.; Welsh, D.A.; Taylor, C.M. Changes in the gut microbial communities following addition of walnuts to the diet. J. Nutr. Biochem. 2017, 48, 94–102. [Google Scholar] [CrossRef]
- Farpour-Lambert, N.; Ells, L.J.; Martinez de Tejada, B.; Scott, C. Obesity and weight gain in Pregnancy and Postpartum: An Evidence Review of Lifestyle Interventions to Inform Maternal and Child Health Policies. Front. Endocrinol. 2018, 9, 546. [Google Scholar] [CrossRef]
- Chu, S.Y.; Kim, S.Y.; Schmid, C.H.; Dietz, P.M.; Callaghan, W.M.; Lau, J.; Curtis, K.M. Maternal obesity and risk of cesarean delivery: A meta-analysis. Obes. Rev. 2007, 8, 385–394. [Google Scholar] [CrossRef]
- Kapinos, K.A.; Yakusheva, O.; Weiss, M. Cesarean deliveries and maternal weight retention. BMC Pregn. Childbirth 2017, 17, 343. [Google Scholar] [CrossRef]
- Knights, D.; Ward, T.L.; McKinlay, C.E.; Miller, H.; Gonzalez, A.; McDonald, D.; Knight, R. Rethinking enterotypes. Cell Host Microbe 2014, 16, 433–437. [Google Scholar] [CrossRef]
- Crusell, M.K.W.; Hansen, T.H.; Nielsen, T.; Allin, K.H.; Rühlemann, M.C.; Damm, P.; Vestergaard, H.; Rørbye, C.; Jørgensen, N.R.; Christiansen, O.B.; et al. Gestational diabetes is associated with change in the gut microbiota composition in third trimester of pregnancy and postpartum. Microbiome 2018, 6, 89. [Google Scholar] [CrossRef]
- Ferrocino, I.; Ponzo, V.; Gambino, R.; Zarovska, A.; Leone, F.; Monzeglio, C.; Goitre, I.; Rosato, R.; Romano, A.; Grassi, G.; et al. Changes in the gut microbiota composition during pregnancy in patients with gestational diabetes mellitus (GDM). Sci. Rep. 2018, 8, 12216. [Google Scholar] [CrossRef]
- Chávez-Carbajal, A.; Nirmalkar, K.; Pérez-Lizaur, A.; Hernández-Quiroz, F.; Ramírez-del-Alto, S.; García-Mena, J.; Hernández-Guerrero, C. Gut Microbiota and Predicted Metabolic Pathways in a Sample of Mexican Women Affected by Obesity and Obesity Plus Metabolic Syndrome. Int. J. Mol. Sci. 2019, 20, 438. [Google Scholar] [CrossRef]
- Liu, H.; Hou, C.; Li, N.; Zhang, X.; Zhang, G.; Yang, F.; Zeng, X.; Liu, Z.; Qiao, S. Microbial and metabolic alterations in gut microbiota of sows during pregnancy and lactation. FASEB J. 2019, 33, 4490–4501. [Google Scholar] [CrossRef]
- Carrothers, J.M.; York, M.A.; Brooker, S.L.; Lackey, K.A.; Williams, J.E.; Shafii, B.; Price, W.J.; Settles, M.L.; McGuire, M.A.; McGuire, M.K. Fecal Microbial Community Structure Is Stable over Time and Related to Variation in Macronutrient and Micronutrient Intakes in Lactating Women. J. Nutr. 2015, 145, 2379–2388. [Google Scholar] [CrossRef] [Green Version]






| Volunteers (%) | Postpartum Periods | |||
|---|---|---|---|---|
| T1 * | T2 | T3 | T4 | |
| Enterotype 1 | 9 | 9 | 9 | 14 |
| Enterotype 2 | 3 | 0 | 0 | 0 |
| Enterotype 3 | 88 | 91 | 91 | 86 |
| UM-0 | 0 | 0 | 0 | 0 |
| UM-A ( | 50 | 50 | 53 | 60 |
| UM-B | 50 | 50 | 47 | 40 |
| Normoweight | 50.0 | 59.4 | 59.4 | 62.5 |
| Overweight | 37.5 | 31.3 | 31.3 | 25.0 |
| Obese | 12.5 | 9.4 | 9.4 | 12.5 |
| All Women (n = 32) | UM-A (n = 16) | UM-B (n = 16) | |
|---|---|---|---|
| BMI (kg/m2) | |||
| T1 * | 25.8 ± 3.5 (19.5–33.3) a | 25.3 ± 2.9 (22.0–30.9) a | 26.3 ± 4.1 (19.5–33.3) a |
| T2 | 25.1 ± 3.8 (19.8–35.8) b | 24.5 ± 2.8 (20.8–29.2) ab | 25.7 ± 4.5 (19.8–35.8) a |
| T3 | 24.9 ± 4.0 (19.8–36.1) b | 24.3 ± 2.9 (20.5–29.5) b | 25.5 ± 4.8 (19.8–36.1) a |
| T4 | 24.3 ± 4.1 (18.7–35.8) c | 23.5 ± 3.3 (18.7–31.1) c | 25.0 ± 4.7 (18.7–35.8) b |
| p-value ⱡ | <0.001 | 0.001 | 0.029 |
| Waist (cm) | |||
| T1 | 92.6 ± 8.0 (76.5–109.0) a | 91.5 ± 6.2 (76.5–99.5) a | 93.6 ± 9.5 (78.7–109.0) a |
| T2 | 89.4 ± 8.3 (68.0–110.0) b | 89.0 ± 7.2 (78.0–101.0) ab | 89.8 ± 9.5 (68.0–110.0) b |
| T3 | 88.0 ± 9.4 (70.0–113.0) bc | 87.0 ± 9.0 (70.0–104.0) b | 89.1 ± 10.0 (76.0–113.0) b |
| T4 | 85.8 ± 10.4 (65.0–115.0) c | 83.0 ± 8.8 (65.0–97.5)c | 88.5 ± 11.3 (72.5–115.0) b |
| p-value ⱡ | <0.001 | 0.001 | 0.021 |
| Hip (cm) | |||
| T1 | 99.9 ± 8.1(86.0–116.0) a | 98.6 ± 6.7 (87.5–110.5) a | 101.2 ± 9.2 (86.0–116.0) a |
| T2 | 95.7 ± 8.7 (78.0–115.0) b | 94.6 ± 9.1 (80.0–109.0) ab | 96.8 ± 8.5 (78.0–115.0) ab |
| T3 | 95.7 ± 9.6 (81.0–123.0) b | 94.0 ± 8.7 (81.0–111.0) b | 97.3 ± 10.5 (83.0–123.0) ab |
| T4 | 94.7 ± 11.1 (78.0–125.0) b | 93.1 ± 10.3 (78.0–111.5) b | 96.5 ± 12.2 (79.0–125.0) b |
| p-value ⱡ | 0.010 | 0.060 | 0.107 |
| Waist/Hip | |||
| T1 | 0.93 ± 0.05 (0.75–1.02) ab | 0.93 ± 0.05 (0.85–1.02) a | 0.93 ± 0.05 (0.75–0.99) a |
| T2 | 0.93 ± 0.05 (0.83–1.03) a | 0.94 ± 0.05 (0.83–1.03) a | 0.93 ± 0.04 (0.83–0.99) a |
| T3 | 0.92 ± 0.04 (0.81–1.00) b | 0.93 ± 0.05 (0.84–1.00) a | 0.92 ± 0.04 (0.81–1.00) a |
| T4 | 0.91 ± 0.06 (0.72–0.99) b | 0.89 ± 0.07 (0.72–0.98) b | 0.93 ± 0.04 (0.87–0.99) a |
| p-value ⱡ | 0.025 | 0.026 | 0.203 |
| Shannon | |||
| T1 | 4.4 ± 0.9 a | 4.3 ± 0.1 a | 4.5 ± 0.1 a |
| T2 | 4.4 ± 0.7 a | 4.3 ± 0.1 a | 4.4 ± 0.1 a |
| T3 | 4.3 ± 0.1 a | 4.2 ± 0.1 a | 4.3 ± 0.2 a |
| T4 | 4.3 ± 0.1 a | 4.2 ± 0.1 a | 4.5 ± 0.1 a |
| p-value ⱡ | 0.607 | 0.749 | 0.659 |
| Chao 1 | |||
| T1 | 2472.2 ± 132.2 a | 2299.6 ± 152.6 a | 2644.8 ± 215.9 a |
| T2 | 2204.5 ± 151.9 a | 2129.1 ± 175.5 a | 2279.8 ± 248.2 ab |
| T3 | 1785.2 ± 154.2 b | 1652.7 ± 178.1 b | 1917.7 ± 251.9 b |
| T4 | 1848.4 ± 110.8 b | 1685.3 ± 127.9 b | 2011.5 ± 181.0 b |
| p-value ⱡ | 0.004 | 0.011 | 0.078 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cortés-Martín, A.; Romo-Vaquero, M.; García-Mantrana, I.; Rodríguez-Varela, A.; Collado, M.C.; Espín, J.C.; Selma, M.V. Urolithin Metabotypes can Anticipate the Different Restoration of the Gut Microbiota and Anthropometric Profiles during the First Year Postpartum. Nutrients 2019, 11, 2079. https://doi.org/10.3390/nu11092079
Cortés-Martín A, Romo-Vaquero M, García-Mantrana I, Rodríguez-Varela A, Collado MC, Espín JC, Selma MV. Urolithin Metabotypes can Anticipate the Different Restoration of the Gut Microbiota and Anthropometric Profiles during the First Year Postpartum. Nutrients. 2019; 11(9):2079. https://doi.org/10.3390/nu11092079
Chicago/Turabian StyleCortés-Martín, Adrián, María Romo-Vaquero, Izaskun García-Mantrana, Ana Rodríguez-Varela, María Carmen Collado, Juan Carlos Espín, and María Victoria Selma. 2019. "Urolithin Metabotypes can Anticipate the Different Restoration of the Gut Microbiota and Anthropometric Profiles during the First Year Postpartum" Nutrients 11, no. 9: 2079. https://doi.org/10.3390/nu11092079