Using the Pleiotropic Characteristics of Curcumin to Validate the Potential Application of a Novel Gene Expression Screening Platform
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
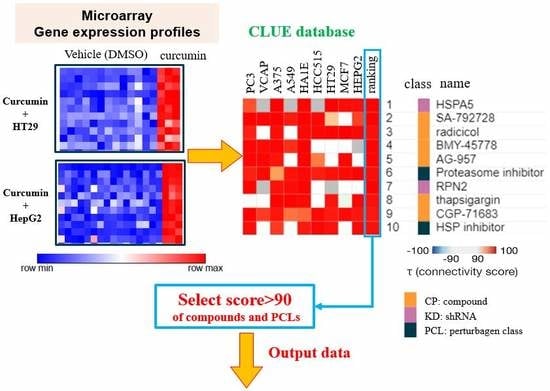
2.2. Determination of Gene Expression Profiles of Curcumin Using the L1000 Microarray
2.3. Cell Culture
2.4. Immunoblotting
2.5. Flow Cytometric Analysis
2.6. Determination of Cell Population Growth
2.7. Statistics
3. Results
3.1. Determining Potential Biological Functions of Curcumin using the C-Map and CLUE Databases
3.2. Validation of Predicted Results Using the Proposed Gene Expression Screening Platform
3.3. Identification of Novel Molecules and Pathways Involved in Curcumin-Suppressed NF-κB
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kumar, G.; Mittal, S.; Sak, K.; Tuli, H.S. Molecular mechanisms underlying chemopreventive potential of curcumin: Current challenges and future perspectives. Life Sci. 2016, 148, 313–328. [Google Scholar] [CrossRef] [PubMed]
- Kunnumakkara, A.B.; Bordoloi, D.; Padmavathi, G.; Monisha, J.; Roy, N.K.; Prasad, S.; Aggarwal, B.B. Curcumin, the golden nutraceutical: multitargeting for multiple chronic diseases. Br. J. Pharmacol. 2017, 174, 1325–1348. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 2017, 171, 1437–1452 e1417. [Google Scholar] [CrossRef] [PubMed]
- Keenan, A.B.; Jenkins, S.L.; Jagodnik, K.M.; Koplev, S.; He, E.; Torre, D.; Wang, Z.; Dohlman, A.B.; Silverstein, M.C.; Lachmann, A.; et al. The library of integrated network-based cellular signatures NIH program: System-level cataloging of human cells response to perturbations. Cell Syst. 2018, 6, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Herwig, R.; Hardt, C.; Lienhard, M.; Kamburov, A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat. Protoc. 2016, 11, 1889–1907. [Google Scholar] [CrossRef] [PubMed]
- Kamburov, A.; Wierling, C.; Lehrach, H.; Herwig, R. ConsensusPathDB--a database for integrating human functional interaction networks. Nucleic Acids Res. 2009, 37, D623–628. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Lee, C.H.; Su, C.L.; Huang, C.W.; Liu, H.S.; Lin, C.N.; Won, S.J. Justicidin A decreases the level of cytosolic Ku70 leading to apoptosis in human colorectal cancer cells. Carcinogenesis 2005, 26, 1716–1730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickman, G.; Julian, L.; Olson, M.F. How apoptotic cells aid in the removal of their own cold dead bodies. Cell Death Differ. 2012, 19, 735–742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, C.L.; Tseng, C.L.; Ramesh, C.; Liu, H.S.; Huang, C.F.; Yao, C.F. Using gene expression database to uncover biology functions of 1,4-disubstituted 1,2,3-triazole analogues synthesized via a copper (I)-catalyzed reaction. Eur. J. Med. Chem. 2017, 132, 90–107. [Google Scholar] [CrossRef]
- Li, M.; Han, S.; Zhang, G.; Wang, Y.; Ji, Z. Antiproliferative activity and apoptosis-inducing mechanism of L-securinine on human breast cancer MCF-7 cells. Pharmazie 2014, 69, 217–223. [Google Scholar] [PubMed]
- Liu, Q.; Kaneko, S.; Yang, L.; Feldman, R.I.; Nicosia, S.V.; Chen, J.; Cheng, J.Q. Aurora-A abrogation of p53 DNA binding and transactivation activity by phosphorylation of serine 215. J. Biol. Chem. 2004, 279, 52175–52182. [Google Scholar] [CrossRef] [PubMed]
- Andresson, T.; Ruderman, J.V. The kinase Eg2 is a component of the Xenopus oocyte progesterone-activated signaling pathway. EMBO J. 1998, 17, 5627–5637. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, S.; Sakashita, G.; Ban, R.; Nagasawa, M.; Matsuzaki, H.; Murata, Y.; Taniguchi, H.; Shima, H.; Furukawa, K.; Urano, T. Phospho-regulation of human protein kinase Aurora-A: analysis using anti-phospho-Thr288 monoclonal antibodies. Oncogene 2006, 25, 7691–7702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, J.; Dong, X.; Zhang, W.; Zeng, X.; Li, Y.; Sun, Y.; Wang, S.; Wang, Z.; Gao, H.; Zhao, W.; et al. Tyrosine kinase inhibitor Thiotanib targets Bcr-Abl and induces apoptosis and autophagy in human chronic myeloid leukemia cells. Appl. Microbiol. Biotechnol. 2014, 98, 9763–9775. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, B.; Li, Q.; Jiang, D.; Yan, S. Anticancer effects of a novel PanRAF inhibitor in a hepatocellular carcinoma cell line. Mol. Med. Rep. 2018, 17, 6185–6193. [Google Scholar]
- Cusack, J.C., Jr.; Liu, R.; Baldwin, A.S., Jr. Inducible chemoresistance to 7-ethyl-10-[4-(1-piperidino)-1-piperidino]-carbonyloxycamptothe cin (CPT-11) in colorectal cancer cells and a xenograft model is overcome by inhibition of nuclear factor-kappaB activation. Cancer Res. 2000, 60, 2323–2330. [Google Scholar]
- Andrews, P.D.; Knatko, E.; Moore, W.J.; Swedlow, J.R. Mitotic mechanics: the auroras come into view. Curr. Opin. Cell Biol. 2003, 15, 672–683. [Google Scholar] [CrossRef]
- Du, J.; Hannon, G.J. Suppression of p160ROCK bypasses cell cycle arrest after Aurora-A/STK15 depletion. Proc. Natl. Acad. Sci. USA 2004, 101, 8975–8980. [Google Scholar] [CrossRef] [Green Version]
- Roth, W.; Reed, J.C. Apoptosis and cancer: When BAX is TRAILing away. Nat. Med. 2002, 8, 216–218. [Google Scholar] [CrossRef]
- Cao, A.; Li, Q.; Yin, P.; Dong, Y.; Shi, H.; Wang, L.; Ji, G.; Xie, J.; Wu, D. Curcumin induces apoptosis in human gastric carcinoma AGS cells and colon carcinoma HT-29 cells through mitochondrial dysfunction and endoplasmic reticulum stress. Apoptosis 2013, 18, 1391–1402. [Google Scholar] [CrossRef] [PubMed]
- Celik, H.; Aydin, T.; Solak, K.; Khalid, S.; Farooqi, A.A. Curcumin on the "flying carpets" to modulate different signal transduction cascades in cancers: Next- generation approach to bridge translational gaps. J. Cell. Biochem. 2018, 119, 4293–4303. [Google Scholar] [CrossRef] [PubMed]
- Salehi, B.; Stojanovic-Radic, Z.; Matejic, J.; Sharifi-Rad, M.; Anil Kumar, N.V.; Martins, N.; Sharifi-Rad, J. The therapeutic potential of curcumin: A review of clinical trials. Eur. J. Med. Chem. 2019, 163, 527–545. [Google Scholar] [CrossRef] [PubMed]
- Asher, G.N.; Spelman, K. Clinical utility of curcumin extract. Altern. Ther. Health Med. 2013, 19, 20–22. [Google Scholar] [PubMed]
- Sharma, R.A.; Steward, W.P.; Gescher, A.J. Pharmacokinetics and pharmacodynamics of curcumin. Adv. Exp. Med. Biol. 2007, 595, 453–470. [Google Scholar] [PubMed]
- Lamb, J. The Connectivity Map: A new tool for biomedical research. Nat. Rev. 2007, 7, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, D.P. Chemotherapeutic potential of curcumin for colorectal cancer. Curr. Pharm. Des. 2002, 8, 1695–1706. [Google Scholar] [CrossRef] [PubMed]
- Dorai, T.; Aggarwal, B.B. Role of chemopreventive agents in cancer therapy. Cancer Lett. 2004, 215, 129–140. [Google Scholar] [CrossRef]
- Patel, B.B.; Majumdar, A.P. Synergistic role of curcumin with current therapeutics in colorectal cancer: Minireview. Nutr. Cancer 2009, 61, 842–846. [Google Scholar] [CrossRef]
- Sharma, R.A.; Gescher, A.J.; Steward, W.P. Curcumin: The story so far. Eur. J. Cancer 2005, 41, 1955–1968. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Shishodia, S. Molecular targets of dietary agents for prevention and therapy of cancer. Biochem. Pharmacol. 2006, 71, 1397–1421. [Google Scholar] [CrossRef] [PubMed]
- Goel, A.; Kunnumakkara, A.B.; Aggarwal, B.B. Curcumin as "Curecumin": From kitchen to clinic. Biochem. Pharmacol. 2008, 75, 787–809. [Google Scholar] [CrossRef] [PubMed]
- Won, S.J.; Wu, H.C.; Lin, K.T.; Yu, C.H.; Chen, Y.T.; Wu, C.S.; Huang, C.Y.F.; Liu, H.S.; Lin, C.N.; Su, C.L. Discovery of molecular mechanisms of lignan justicidin A using L1000 gene expression profiles and the Library of Integrated Network-based Cellular Signatures database. J. Funct. Foods 2015, 16, 81–93. [Google Scholar] [CrossRef]
- Rice, N.R.; Ernst, M.K. In vivo control of NF-kappa B activation by I kappa B alpha. EMBO J. 1993, 12, 4685–4695. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Hawke, N.; Baldwin, A.S. NF-kappaB and IKK as therapeutic targets in cancer. Cell Death Differ. 2006, 13, 738–747. [Google Scholar] [CrossRef] [PubMed]
- Düwel, M.; Welteke, V.; Oeckinghaus, A.; Baens, M.; Kloo, B.; Ferch, U.; Darnay, B.G.; Ruland, J.; Marynen, P.; Krappmann, D. A20 negatively regulates T cell receptor signaling to NF-kappaB by cleaving Malt1 ubiquitin chains. J. Immunol. 2009, 182, 7718–7728. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.T.; Wu, A.T.; Chang, P.M.; Chen, K.Y.; Yang, C.N.; Yang, S.C.; Ho, C.C.; Chen, C.C.; Kuo, Y.L.; Lee, P.Y.; et al. Trifluoperazine, an antipsychotic agent, inhibits cancer stem cell growth and overcomes drug resistance of lung cancer. Am. J. Respir. Crit. Care Med. 2012, 186, 1180–1188. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.T.; Su, C.L.; Huang, C.Y.; Lin, J.K.; Lee, W.H.; Chang, P.M.; Kuo, Y.L.; Liu, Y.W.; Wang, L.S.; Wu, C.H.; et al. A preclinical evaluation of antimycin a as a potential antilung cancer stem cell agent. Evid. Based Complement. Altern. Med. 2013, 2013, 910451. [Google Scholar] [CrossRef]
- Cheng, H.W.; Liang, Y.H.; Kuo, Y.L.; Chuu, C.P.; Lin, C.Y.; Lee, M.H.; Wu, A.T.; Yeh, C.T.; Chen, E.I.; Whang-Peng, J.; et al. Identification of thioridazine, an antipsychotic drug, as an antiglioblastoma and anticancer stem cell agent using public gene expression data. Cell Death Dis. 2015, 6, e1753. [Google Scholar] [CrossRef]
- Kuo, Y.L.; Chen, C.H.; Chuang, T.H.; Hua, W.K.; Lin, W.J.; Hsu, W.H.; Chang, P.M.; Hsu, S.L.; Huang, T.H.; Kao, C.Y.; et al. Gene expression profiling and pathway network analysis predicts a novel antitumor function for a botanical-derived drug, PG2. Evid. Based Complement. Altern. Med. 2015, 2015, 917345. [Google Scholar] [CrossRef]








© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, S.-C.; Hsu, H.-W.; Chuang, K.-L.; Huang, Z.-Y.; Lin, K.-T.; Hsu, W.-H.; Chang, K.-H.; Huang, C.-Y.F.; Su, C.-L. Using the Pleiotropic Characteristics of Curcumin to Validate the Potential Application of a Novel Gene Expression Screening Platform. Nutrients 2019, 11, 1397. https://doi.org/10.3390/nu11061397
Liao S-C, Hsu H-W, Chuang K-L, Huang Z-Y, Lin K-T, Hsu W-H, Chang K-H, Huang C-YF, Su C-L. Using the Pleiotropic Characteristics of Curcumin to Validate the Potential Application of a Novel Gene Expression Screening Platform. Nutrients. 2019; 11(6):1397. https://doi.org/10.3390/nu11061397
Chicago/Turabian StyleLiao, Se-Chun, Hsiu-Wen Hsu, Kun-Lin Chuang, Zi-Yi Huang, Kuan-Ting Lin, Wei-Hsiang Hsu, Kai-Hsun Chang, Chi-Yin F. Huang, and Chun-Li Su. 2019. "Using the Pleiotropic Characteristics of Curcumin to Validate the Potential Application of a Novel Gene Expression Screening Platform" Nutrients 11, no. 6: 1397. https://doi.org/10.3390/nu11061397