Key Genes of Lipid Metabolism and WNT-Signaling Are Downregulated in Subcutaneous Adipose Tissue with Moderate Weight Loss
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. Blood-Based Biomarkers
2.3. Adipose Tissue Biopsies
2.4. mRNA Extraction and Microarray Analyses
2.5. Reverse Transcription and Quantitative Polymerase Chain Reaction (RT-qPCR)
2.6. Literature Overview
2.7. Statistical Analyses
3. Results
3.1. Characteristics of the Study Cohort
3.2. Microarray Gene Expression by Weight Loss Groups
3.3. Validation of Gene Expression Levels for SCD, SFRP2, and HILPDA
3.4. Circulating Levels of HILPDA and SFRP2
3.5. Correlation of Adipose-Tissue Gene Expression with BMI and Metabolic Biomarkers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Renehan, A.G.; Zwahlen, M.; Egger, M. Adiposity and cancer risk: New mechanistic insights from epidemiology. Nat. Rev. Cancer 2015, 15, 484–498. [Google Scholar] [CrossRef]
- Calle, E.E.; Kaaks, R. Overweight, obesity and cancer: Epidemiological evidence and proposed mechanisms. Nat. Rev. Cancer 2004, 4, 579–591. [Google Scholar] [CrossRef]
- Tchernof, A.; Despres, J.P. Pathophysiology of human visceral obesity: An update. Physiol. Rev. 2013, 93, 359–404. [Google Scholar] [CrossRef]
- Tilg, H.; Moschen, A.R. Adipocytokines: Mediators linking adipose tissue, inflammation and immunity. Nat. Rev. Immunol. 2006, 6, 772–783. [Google Scholar] [CrossRef]
- Jensen, M.D.; Ryan, D.H.; Apovian, C.M.; Ard, J.D.; Comuzzie, A.G.; Donato, K.A.; Hu, F.B.; Hubbard, V.S.; Jakicic, J.M.; Kushner, R.F.; et al. 2013 AHA/ACC/TOS guideline for the management of overweight and obesity in adults. Circulation 2013, 129, S102–S138. [Google Scholar] [CrossRef]
- Magkos, F.; Fraterrigo, G.; Yoshino, J.; Luecking, C.; Kirbach, K.; Kelly, S.C.; de Las Fuentes, L.; He, S.; Okunade, A.L.; Patterson, B.W.; et al. Effects of moderate and subsequent progressive weight loss on metabolic function and adipose tissue biology in humans with obesity. Cell Metab. 2016, 23, 591–601. [Google Scholar] [CrossRef]
- Mutch, D.M.; Pers, T.H.; Temanni, M.R.; Pelloux, V.; Marquez-Quinones, A.; Holst, C.; Martinez, J.A.; Babalis, D.; van Baak, M.A.; Handjieva-Darlenska, T.; et al. A distinct adipose tissue gene expression response to caloric restriction predicts 6-mo weight maintenance in obese subjects. Am. J. Clin. Nutr. 2011, 94, 1399–1409. [Google Scholar] [CrossRef]
- Klimcakova, E.; Roussel, B.; Marquez-Quinones, A.; Kovacova, Z.; Kovacikova, M.; Combes, M.; Siklova-Vitkova, M.; Hejnova, J.; Sramkova, P.; Bouloumie, A.; et al. Worsening of obesity and metabolic status yields similar molecular adaptations in human subcutaneous and visceral adipose tissue: Decreased metabolism and increased immune response. J. Clin. Endocrinol. Metab. 2011, 96, E73–E82. [Google Scholar] [CrossRef]
- Moreno-Navarrete, J.M.; Petrov, P.; Serrano, M.; Ortega, F.; Garcia-Ruiz, E.; Oliver, P.; Ribot, J.; Ricart, W.; Palou, A.; Bonet, M.L.; et al. Decreased rb1 mrna, protein, and activity reflect obesity-induced altered adipogenic capacity in human adipose tissue. Diabetes 2013, 62, 1923–1931. [Google Scholar] [CrossRef]
- Mardinoglu, A.; Heiker, J.T.; Gartner, D.; Bjornson, E.; Schon, M.R.; Flehmig, G.; Kloting, N.; Krohn, K.; Fasshauer, M.; Stumvoll, M.; et al. Extensive weight loss reveals distinct gene expression changes in human subcutaneous and visceral adipose tissue. Sci. Rep. 2015, 5, 14841. [Google Scholar] [CrossRef]
- Liu, Y.; Aron-Wisnewsky, J.; Marcelin, G.; Genser, L.; Le Naour, G.; Torcivia, A.; Bauvois, B.; Bouchet, S.; Pelloux, V.; Sasso, M.; et al. Accumulation and changes in composition of collagens in subcutaneous adipose tissue after bariatric surgery. J. Clin. Endocrinol. Metab. 2016, 101, 293–304. [Google Scholar] [CrossRef]
- Schübel, R.; Nattenmüller, J.; Sookthai, D.; Nonnenmacher, T.; Graf, M.E.; Riedl, L.; Schlett, C.L.; von Stackelberg, O.; Johnson, T.; Nabers, D.; et al. Effects of intermittent and continuous calorie restriction on body weight and metabolism over 50 weeks: A randomized controlled trial. Am. J. Clin. Nutr. 2018, 108, 933–945. [Google Scholar] [CrossRef]
- Schübel, R.; Graf, M.E.; Nattenmüller, J.; Nabers, D.; Sookthai, D.; Gruner, L.F.; Johnson, T.; Schlett, C.L.; von Stackelberg, O.; Kirsten, R.; et al. The effects of intermittent calorie restriction on metabolic health: Rationale and study design of the helena trial. Contemp. Clin. Trials 2016, 51, 28–33. [Google Scholar] [CrossRef]
- Oberritter, H.; Schabethal, K.; von Ruesten, A.; Boeing, H. The dge nutrition circle—Presentation and basis of the food-related recommendations from the german nutrition society (dge). Ernahrungs Umschau 2013, 60, 24–29. [Google Scholar]
- Pakiet, A.; Kobiela, J.; Stepnowski, P.; Sledzinski, T.; Mika, A. Changes in lipids composition and metabolism in colorectal cancer: A review. Lipids Health Dis. 2019, 18, 29. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, Q.; Zhou, D.; Huang, C.; Meng, X.; Li, J. Secreted frizzled-related protein 2-mediated cancer events: Friend or foe? Pharmacol. Rep. 2017, 69, 403–408. [Google Scholar] [CrossRef]
- DiStefano, M.T.; Roth Flach, R.J.; Senol-Cosar, O.; Danai, L.V.; Virbasius, J.V.; Nicoloro, S.M.; Straubhaar, J.; Dagdeviren, S.; Wabitsch, M.; Gupta, O.T.; et al. Adipocyte-specific hypoxia-inducible gene 2 promotes fat deposition and diet-induced insulin resistance. Mol. Metab. 2016, 5, 1149–1161. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The miqe guidelines: Minimum information for publication of quantitative real-time pcr experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.; Wakem, M.; Dijkman, G.; Alsarraj, M.; Nguyen, M. A practical approach to rt-qpcr-publishing data that conform to the miqe guidelines. Methods 2010, 50, S1–S5. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Troyanskaya, O.; Cantor, M.; Sherlock, G.; Brown, P.; Hastie, T.; Tibshirani, R.; Botstein, D.; Altman, R.B. Missing value estimation methods for DNA microarrays. Bioinformatics 2001, 17, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Christodoulides, C.; Lagathu, C.; Sethi, J.K.; Vidal-Puig, A. Adipogenesis and wnt signalling. Trends Endocrinol. Metab. 2009, 20, 16–24. [Google Scholar] [CrossRef]
- Park, J.R.; Jung, J.W.; Lee, Y.S.; Kang, K.S. The roles of wnt antagonists dkk1 and sfrp4 during adipogenesis of human adipose tissue-derived mesenchymal stem cells. Cell Prolif. 2008, 41, 859–874. [Google Scholar] [CrossRef] [PubMed]
- Lagathu, C.; Christodoulides, C.; Tan, C.Y.; Virtue, S.; Laudes, M.; Campbell, M.; Ishikawa, K.; Ortega, F.; Tinahones, F.J.; Fernandez-Real, J.M.; et al. Secreted frizzled-related protein 1 regulates adipose tissue expansion and is dysregulated in severe obesity. Int. J. Obes. 2010, 34, 1695–1705. [Google Scholar] [CrossRef] [PubMed]
- Cianferotti, L.; Demay, M.B. Vdr-mediated inhibition of dkk1 and sfrp2 suppresses adipogenic differentiation of murine bone marrow stromal cells. J. Cell. Biochem. 2007, 101, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Dahlman, I.; Mejhert, N.; Linder, K.; Agustsson, T.; Mutch, D.M.; Kulyte, A.; Isaksson, B.; Permert, J.; Petrovic, N.; Nedergaard, J.; et al. Adipose tissue pathways involved in weight loss of cancer cachexia. Br. J. Cancer 2010, 102, 1541–1548. [Google Scholar] [CrossRef] [PubMed]
- Johansson, L.E.; Danielsson, A.P.; Parikh, H.; Klintenberg, M.; Norstrom, F.; Groop, L.; Ridderstrale, M. Differential gene expression in adipose tissue from obese human subjects during weight loss and weight maintenance. Am. J. Clin. Nutr. 2012, 96, 196–207. [Google Scholar] [CrossRef]
- Armenise, C.; Lefebvre, G.; Carayol, J.; Bonnel, S.; Bolton, J.; Di Cara, A.; Gheldof, N.; Descombes, P.; Langin, D.; Saris, W.H.; et al. Transcriptome profiling from adipose tissue during a low-calorie diet reveals predictors of weight and glycemic outcomes in obese, nondiabetic subjects. Am. J. Clin. Nutr. 2017, 106, 736–746. [Google Scholar] [CrossRef]
- Ehrlund, A.; Mejhert, N.; Lorente-Cebrian, S.; Astrom, G.; Dahlman, I.; Laurencikiene, J.; Ryden, M. Characterization of the wnt inhibitors secreted frizzled-related proteins (sfrps) in human adipose tissue. J. Clin. Endocrinol. Metab. 2013, 98, E503–E508. [Google Scholar] [CrossRef]
- Crowley, R.K.; O’Reilly, M.W.; Bujalska, I.J.; Hassan-Smith, Z.K.; Hazlehurst, J.M.; Foucault, D.R.; Stewart, P.M.; Tomlinson, J.W. Sfrp2 is associated with increased adiposity and vegf expression. PLoS ONE 2016, 11, e0163777. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Webster, M.R.; Marchbank, K.; Behera, R.; Ndoye, A.; Kugel, C.H., 3rd; Dang, V.M.; Appleton, J.; O’Connell, M.P.; Cheng, P.; et al. Sfrp2 in the aged microenvironment drives melanoma metastasis and therapy resistance. Nature 2016, 532, 250–254. [Google Scholar] [CrossRef] [PubMed]
- DiStefano, M.T.; Danai, L.V.; Roth Flach, R.J.; Chawla, A.; Pedersen, D.J.; Guilherme, A.; Czech, M.P. The lipid droplet protein hypoxia-inducible gene 2 promotes hepatic triglyceride deposition by inhibiting lipolysis. J. Biol. Chem. 2015, 290, 15175–15184. [Google Scholar] [CrossRef] [PubMed]
- Mattijssen, F.; Georgiadi, A.; Andasarie, T.; Szalowska, E.; Zota, A.; Krones-Herzig, A.; Heier, C.; Ratman, D.; De Bosscher, K.; Qi, L.; et al. Hypoxia-inducible lipid droplet-associated (hilpda) is a novel peroxisome proliferator-activated receptor (ppar) target involved in hepatic triglyceride secretion. J. Biol. Chem. 2014, 289, 19279–19293. [Google Scholar] [CrossRef] [PubMed]
- Denko, N.; Schindler, C.; Koong, A.; Laderoute, K.; Green, C.; Giaccia, A. Epigenetic regulation of gene expression in cervical cancer cells by the tumor microenvironment. Clin. Cancer Res. 2000, 6, 480–487. [Google Scholar]
- Gimm, T.; Wiese, M.; Teschemacher, B.; Deggerich, A.; Schodel, J.; Knaup, K.X.; Hackenbeck, T.; Hellerbrand, C.; Amann, K.; Wiesener, M.S.; et al. Hypoxia-inducible protein 2 is a novel lipid droplet protein and a specific target gene of hypoxia-inducible factor-1. FASEB J. 2010, 24, 4443–4458. [Google Scholar] [CrossRef]
- Applebaum, M.A.; Jha, A.R.; Kao, C.; Hernandez, K.M.; DeWane, G.; Salwen, H.R.; Chlenski, A.; Dobratic, M.; Mariani, C.J.; Godley, L.A.; et al. Integrative genomics reveals hypoxia inducible genes that are associated with a poor prognosis in neuroblastoma patients. Oncotarget 2016, 7, 76816–76826. [Google Scholar] [CrossRef]
- Franck, N.; Gummesson, A.; Jernas, M.; Glad, C.; Svensson, P.A.; Guillot, G.; Rudemo, M.; Nystrom, F.H.; Carlsson, L.M.; Olsson, B. Identification of adipocyte genes regulated by caloric intake. J. Clin. Endocrinol. Metab. 2011, 96, E413–E418. [Google Scholar] [CrossRef]
- Capel, F.; Viguerie, N.; Vega, N.; Dejean, S.; Arner, P.; Klimcakova, E.; Martinez, J.A.; Saris, W.H.; Holst, C.; Taylor, M.; et al. Contribution of energy restriction and macronutrient composition to changes in adipose tissue gene expression during dietary weight-loss programs in obese women. J. Clin. Endocrinol. Metab. 2008, 93, 4315–4322. [Google Scholar] [CrossRef]
- Poudyal, H.; Brown, L. Stearoyl-coa desaturase: A vital checkpoint in the development and progression of obesity. Endocr. Metab. Immune Disorders Drug Targets 2011, 11, 217–231. [Google Scholar] [CrossRef]
- Ntambi, J.M.; Miyazaki, M. Regulation of stearoyl-coa desaturases and role in metabolism. Prog. Lipid Res. 2004, 43, 91–104. [Google Scholar] [CrossRef]
- Kroger, J.; Schulze, M.B. Recent insights into the relation of delta5 desaturase and delta6 desaturase activity to the development of type 2 diabetes. Curr. Opin. Lipidol. 2012, 23, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Tosi, F.; Sartori, F.; Guarini, P.; Olivieri, O.; Martinelli, N. Delta-5 and delta-6 desaturases: Crucial enzymes in polyunsaturated fatty acid-related pathways with pleiotropic influences in health and disease. Adv. Exp. Med. Biol. 2014, 824, 61–81. [Google Scholar]
- Inadera, H. The usefulness of circulating adipokine levels for the assessment of obesity-related health problems. Int. J. Med. Sci. 2008, 5, 248–262. [Google Scholar] [CrossRef]
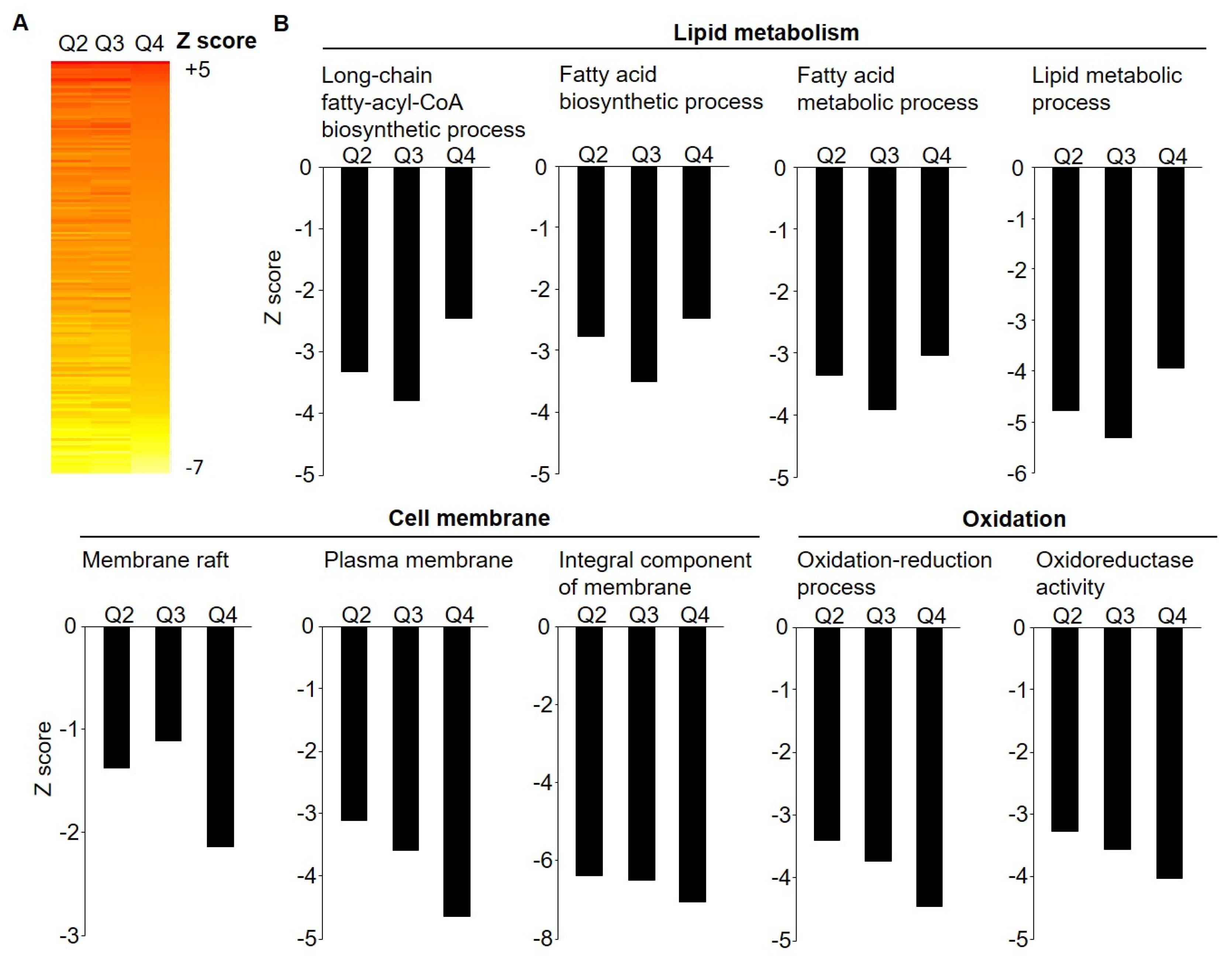
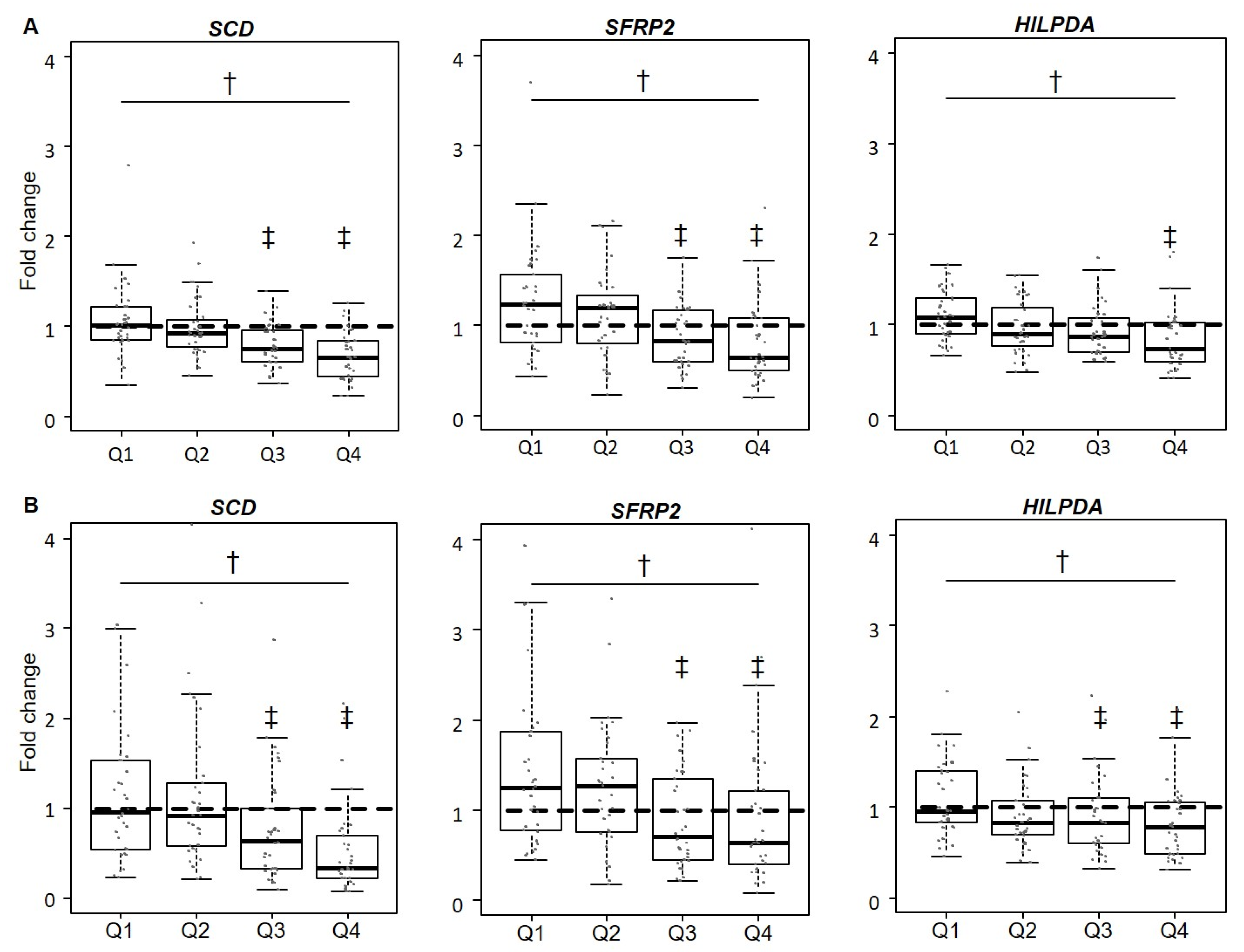
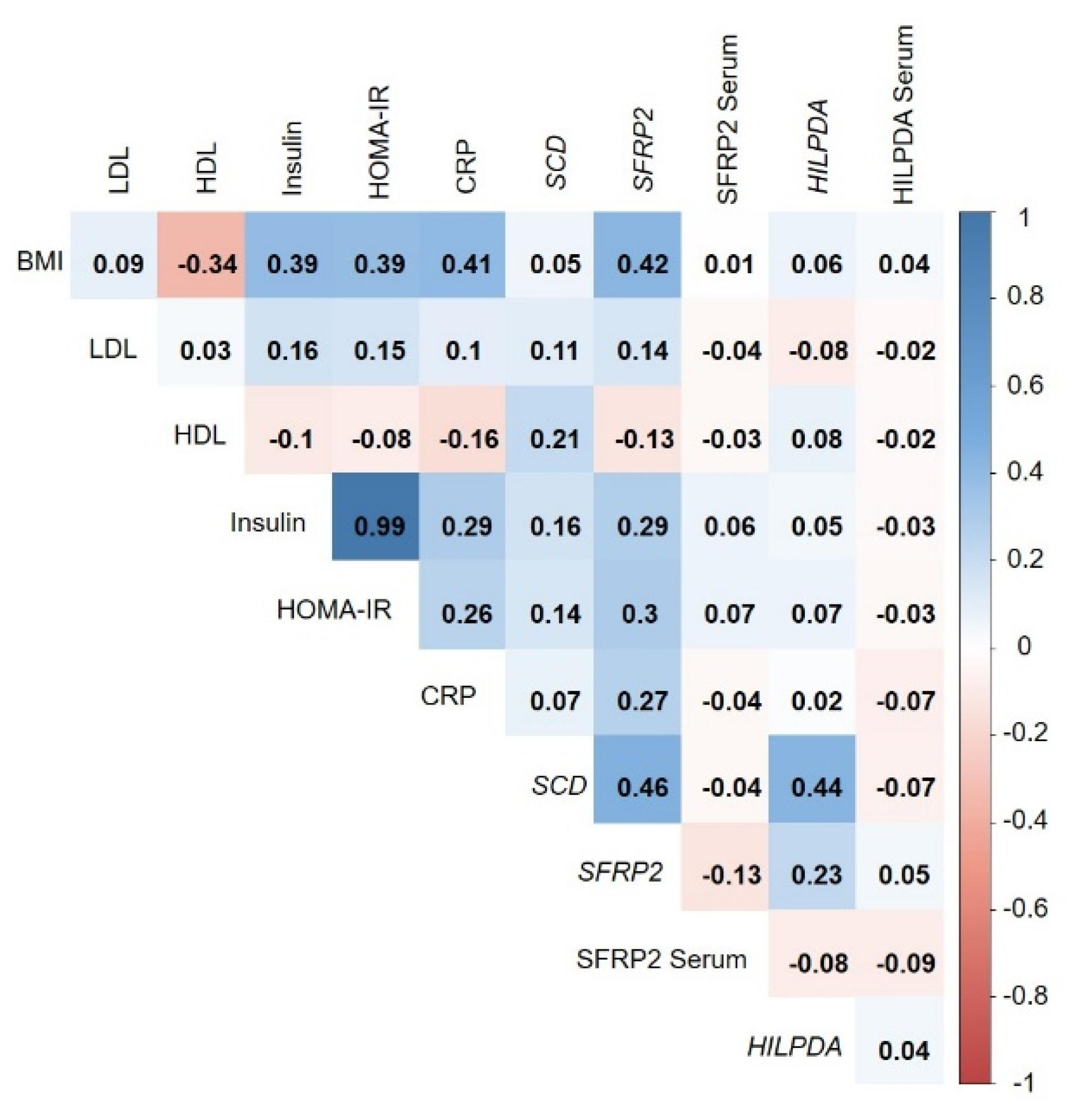
| Quartile 1 | Quartile 2 | Quartile 3 | Quartile 4 | |
|---|---|---|---|---|
| (n = 35) | (n = 34) | (n = 35) | (n = 34) | |
| Women, n (%) | 17 (48.6) | 18 (52.9) | 14 (40.0) | 18 (52.9) |
| Age (years) | 47.2 ± 8.3 | 52.0 ± 7.4 | 50.6± 8.7 | 50.6 ± 6.5 |
| Weight (kg) | 94.1 ± 14.4 | 93.2 ± 15.9 | 94.4 ± 13.7 | 94.5 ± 16.3 |
| Height (cm) | 173.4 ± 8.0 | 173.7 ± 10.9 | 173.6 ± 9.4 | 172.5 ± 10.3 |
| BMI (kg/m2) | 31.5 ± 3.7 | 30.8 ± 3.4 | 31.3 ± 3.8 | 31.7 ± 4.2 |
| Education level, n (%) 2 | ||||
| Primary school | 4 (12.1) | 2 (5.9) | 5 (14.7) | 2 (5.7) |
| Secondary school | 14 (42.4) | 9 (26.4) | 6 (17.7) | 8 (22.9) |
| Higher education | 15 (45.5) | 23 (67.7) | 23 (67.6) | 25 (71.4) |
| Glucose (mg/dL) | 91.5 ± 7.9 | 95.1 ± 6.6 | 93.0 ± 6.6 | 93.0± 7.8 |
| Insulin (mU/L) | 11.0 ± 5.3 | 10.6 ± 5.0 | 13.0 ± 7.4 | 13.6 ± 7.0 |
| HOMA-IR | 2.5 ± 1.3 | 2.5 ± 1.2 | 3.0 ± 1.8 | 3.1 ± 1.6 |
| Cholesterol (mg/dL) | 212.8 ± 35.2 | 199.7 ± 31.7 | 214.9 ± 36.8 | 202.9 ± 34.9 |
| HDL-cholesterol (mg/dL) | 53.8 ± 15.9 | 51.8 ± 13.5 | 57.6 ± 13.7 | 52.9 ± 15.1 |
| LDL-cholesterol (mg/dL) | 128.8 ± 24.8 | 120.8 ± 25.1 | 129.2 ± 26.8 | 128.4 ± 29.8 |
| Address | Gene | Description | Highest vs. Lowest | ||
|---|---|---|---|---|---|
| ID | Symbol | Weight Loss Quartile | |||
| Log2 FC 1 | p Value 1 | ptrend Value 2 | |||
| Downregulated genes | |||||
| 2360020 | FADS1 | Fatty acid desaturase 1 | −1.09 | <0.001 | <0.001 |
| 4850195 | SCD | Stearoyl-CoA desaturase | −1.00 | <0.001 | <0.001 |
| 2140128 | SCD | Stearoyl-CoA desaturase | −0.69 | <0.001 | <0.001 |
| 3060639 | SFRP2 | Secreted frizzled-related protein 2 | −0.65 | 0.006 | 0.004 |
| 6200253 | THBS4 | Thrombospondin 4 | −0.58 | 0.019 | 0.026 |
| 7040372 | GPAM | Glycerol-3-phosphate acyltransferase | −0.54 | <0.001 | 0.001 |
| 7330544 | ALDOC | Aldolase C, fructose-bisphosphate | −0.52 | <0.001 | 0.001 |
| 3840026 | GPAM | Glycerol-3-phosphate acyltransferase | −0.51 | <0.001 | <0.001 |
| 5090026 | COL15A1 | Collagen, type XV, alpha 1 | −0.50 | <0.001 | <0.001 |
| 5550292 | KLB | Klotho beta | −0.49 | <0.001 | <0.001 |
| 240400 | PMEPA1 | Prostate transmembrane protein | −0.48 | 0.002 | 0.001 |
| 4010709 | NNAT | Neuronatin | −0.47 | 0.008 | 0.005 |
| 3390326 | ME1 | Malic enzyme 1, NADP(+)-dependent | −0.47 | <0.001 | <0.001 |
| 5550379 | CAV1 | Caveolin 1 | −0.47 | 0.0041 | 0.017 |
| 7650053 | ECHDC1 | Enoyl CoA hydratase domain containing 1 | −0.46 | <0.001 | <0.001 |
| 450292 | TUBB2A | Tubulin | −0.46 | 0.008 | 0.011 |
| 7320441 | HILPDA | Hypoxia inducible lipid droplet-associated | −0.45 | <0.001 | <0.001 |
| 4480341 | DHCR24 | 24-dehydrocholesterol reductase | −0.45 | <0.001 | <0.001 |
| 2370041 | LRRN3 | Leucine rich repeat neuronal 3 | −0.45 | <0.001 | 0.0012 |
| 3450537 | DGAT2 | Diacylglycerol O-acyltransferase 2 | −0.44 | 0.015 | 0.016 |
| 7610128 | KANK4 | KN motif and ankyrin repeat domains 4 | −0.44 | 0.032 | 0.012 |
| 2480338 | DBI | Diazepam binding inhibitor | −0.44 | <0.001 | <0.001 |
| 730040 | LAMB3 | Laminin, beta 3 | −0.43 | <0.001 | <0.001 |
| 3830041 | PMEPA1 | Prostate transmembrane protein | −0.43 | 0.0061 | 0.0043 |
| 360192 | INSIG1 | Insulin induced gene 1 | −0.43 | <0.001 | <0.001 |
| 3180048 | IDH1 | Isocitrate dehydrogenase 1 | −0.42 | <0.001 | <0.001 |
| 2230538 | LRRN3 | Leucine rich repeat neuronal 3 | −0.41 | 0.002 | 0.003 |
| 1710484 | FMOD | Fibromodulin | −0.41 | 0.002 | 0.002 |
| 3390343 | SREBF1 | Sterol regulatory element binding | −0.41 | 0.029 | 0.022 |
| 3170594 | MAL2 | Mal, T-cell differentiation protein 2 | −0.41 | 0.013 | 0.020 |
| 5310634 | FASN | Fatty acid synthase | −0.41 | 0.013 | 0.0118 |
| 520474 | TENM4 | Teneurin transmembrane protein 4 | −0.40 | <0.001 | <0.001 |
| Upregulated genes | |||||
| 6520040 | C6 | Complement component 6 | 0.66 | <0.001 | <0.001 |
| 6280370 | LOC646688 | Predicted: misc_RNA | 0.51 | <0.001 | <0.001 |
| 2140278 | CIDEA | Cell death-inducing DFFA-like effector a | 0.47 | 0.015 | <0.001 |
| 2490612 | ADH1B | Alcohol dehydrogenase IB | 0.45 | <0.001 | <0.001 |
| 7560543 | MOCS1 | Molybdenum cofactor synthesis 1 | 0.44 | <0.001 | <0.001 |
| 10048 | CIDEA | Cell death-inducing DFFA-like effector a | 0.44 | 0.012 | <0.001 |
| 5490019 | GPX3 | Glutathione peroxidase 3 | 0.41 | 0.003 | <0.001 |
| 6270372 | EGFLAM | EGF-like, fibronectin type III and laminin G domains | 0.40 | 0.012 | <0.001 |
| n | Baseline | n | Week 12 | Relative | p-Value 1 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD | 95% CI | Mean ± SD | 95% CI | Change | All | Q1 vs. Q4 | |||
| HILPDA (µg/mL) | |||||||||
| Quartile 1 | 33 | 51.8± 26.5 | (44.0, 59.6) | 32 | 52.8 ± 24.2 | (45.5, 60.1) | 6.8 ± 4.8 | 0.78 | 0.84 |
| Quartile 2 | 34 | 55.6 ± 25.3 | (48.2, 62.9) | 33 | 53.8 ± 23.9 | (46.8, 60.9) | 0.5 ± 7.4 | ||
| Quartile 3 | 32 | 49.0 ± 28.5 | (40.5, 57.6) | 34 | 48.9 ± 27.4 | (41.0, 56.9) | 4.3 ± 6.3 | ||
| Quartile 4 | 35 | 53.2 ± 29.0 | (44.9, 61.5) | 34 | 54.4 ± 28.5 | (46.2, 62.7) | 5.8 ± 9.2 | ||
| SFRP2 (ng/mL) | |||||||||
| Quartile 1 | 32 | 36.5 ± 29.5 | (27.6, 45.3) | 27 | 38.1 ± 34.1 | (26.9, 49.3) | 2.8 ± 7.1 | 0.36 | 0.31 |
| Quartile 2 | 31 | 45.6 ± 26.5 | (37.5, 53.7) | 30 | 39.3 ± 19.5 | (33.3, 45.4) | −3.0 ± 5.3 | ||
| Quartile 3 | 31 | 29.3 ± 13.6 | (25.2, 33.4) | 32 | 33.2 ± 15.3 | (28.7, 37.8) | 11.2 ± 8.9 | ||
| Quartile 4 | 33 | 45.0 ± 27.5 | (36.9, 53.1) | 31 | 46.1 ± 26.1 | (38.2, 54.0) | −3.6 ± 8.1 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schübel, R.; Sookthai, D.; Greimel, J.; Johnson, T.S.; Grafetstätter, M.E.; Kirsten, R.; Kratz, M.; Ulrich, C.M.; Kaaks, R.; Kühn, T. Key Genes of Lipid Metabolism and WNT-Signaling Are Downregulated in Subcutaneous Adipose Tissue with Moderate Weight Loss. Nutrients 2019, 11, 639. https://doi.org/10.3390/nu11030639
Schübel R, Sookthai D, Greimel J, Johnson TS, Grafetstätter ME, Kirsten R, Kratz M, Ulrich CM, Kaaks R, Kühn T. Key Genes of Lipid Metabolism and WNT-Signaling Are Downregulated in Subcutaneous Adipose Tissue with Moderate Weight Loss. Nutrients. 2019; 11(3):639. https://doi.org/10.3390/nu11030639
Chicago/Turabian StyleSchübel, Ruth, Disorn Sookthai, Judith Greimel, Theron S. Johnson, Mirja E. Grafetstätter, Romy Kirsten, Mario Kratz, Cornelia M. Ulrich, Rudolf Kaaks, and Tilman Kühn. 2019. "Key Genes of Lipid Metabolism and WNT-Signaling Are Downregulated in Subcutaneous Adipose Tissue with Moderate Weight Loss" Nutrients 11, no. 3: 639. https://doi.org/10.3390/nu11030639
APA StyleSchübel, R., Sookthai, D., Greimel, J., Johnson, T. S., Grafetstätter, M. E., Kirsten, R., Kratz, M., Ulrich, C. M., Kaaks, R., & Kühn, T. (2019). Key Genes of Lipid Metabolism and WNT-Signaling Are Downregulated in Subcutaneous Adipose Tissue with Moderate Weight Loss. Nutrients, 11(3), 639. https://doi.org/10.3390/nu11030639





