Proteomics Analysis Reveals the Implications of Cytoskeleton and Mitochondria in the Response of the Rat Brain to Starvation
Abstract
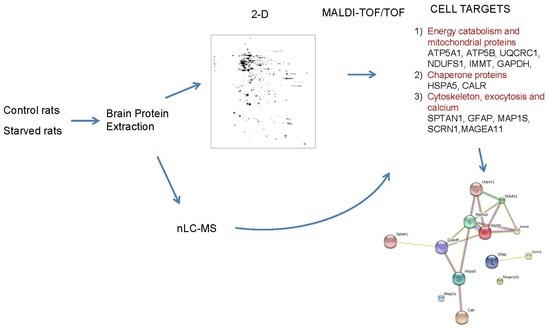
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Drugs
2.2. Animals and Experimental Design
2.3. Protein Extraction for 2-D
2.4. 2-D
2.5. Quantitative Analysis of Gel Images and Statistical Analysis
2.6. Protein Digestion and MS Analysis
2.7. Database Searching
2.8. nLC-MS Proteomics Procedure
2.8.1. Sample Preparation
2.8.2. Protein Digestion
2.8.3. nLC-MS2 Analysis
2.8.4. Data Analysis
2.8.5. Quantitative First-Mass MS1 Data Analysis in Skyline Software
3. Results
4. Discussion
Author Contributions
Acknowledgements
Conflicts of Interest
Abbreviations
| 2-D | two-dimensional gel electrophoresis |
| MS | mass spectrometry |
| MALDI-TOF/TO | Matrix-assisted laser desorption-ionization time of flight/time of flight |
| nLC-MS | nanoliquid chromatography and MS analysis |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| ATP5A1 | α subunit of ATP synthase |
| ATP5B | β subunit of ATP synthase |
| UQCRC1 | subunit 1 of the cytochrome b-c1 complex |
| NDUFS1 | subunit of 75 kDa of NADH-ubiquinone oxidoreductase |
| IMMT | MIC60 subunit of the MICOS (internal mitochondrial membrane protein) |
| HSPA5 | including 78 kDa glucose-regulated protein |
| CALR | calreticulin |
| SPTAN1 | α-chain of non-erythrocytic spectine 1 |
| GFAP | glial fibrillary acidic protein |
| MAP1S | 1S microtubule-associated protein |
| SCRN1 | Secernin-1 |
| MAGEA11 | melanoma-associated antigen |
References
- Sokolović, A.; Roomen, C.P.; Ottenhoff, R.; Scheij, S.; Hiralall, J.K.; Claessen, N.; Aten, J.; Oude Elferink, R.P.J.; Groen, A.K.; Sokolović, M. Fasting reduces liver fibrosis in a mouse model for chronic cholangiopathies. Biochim. Biophys. Acta 2013, 1832, 1482–1491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hakvoort, T.B.; Moerland, P.D.; Frijters, R.; Sokolovic, A.; Labruyère, W.T.; Vermeulen, J.L.; Ver Loren van Themaat, E.; Breit, T.M.; Wittink, F.R.A.; van Kampen, A.H.C.; et al. Interorgan coordination of the murine adaptive response to fasting. J. Biol. Chem. 2011, 286, 16332–16343. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, P.N.; Gasperowicz, M.; Barbeito-Andrés, J.; Klenin, N.; Cross, J.C.; Hallgrimsson, B. Chronic protein restriction in mice impacts placental function and maternal body weight before fetal growth. PLoS ONE 2016, 11, e0152227. [Google Scholar] [CrossRef] [PubMed]
- Shimizu-Albergine, M.; Ippolito, D.L.; Beavo, J.A. Downregulation of fasting-induced cAMP response element-mediated gene induction by leptin in neuropeptide Y neurons of the arcuate nucleus. J. Neurosci. 2001, 21, 1238–1246. [Google Scholar] [CrossRef] [PubMed]
- Owen, O.; Morgan, A.; Kemp, H.; Sullivan, J.; Herrera, M.; Cahill, G.F.J. Brain metabolism during fasting. J. Clin. Investig. 1967, 46, 1589–1595. [Google Scholar] [CrossRef] [PubMed]
- LaManna, J.C.; Salem, N.; Puchowicz, M.; Erokwu, B.; Koppaka, S.; Flask, C.; Lee, Z. Ketones suppress brain glucose consumption. Adv. Exp. Med. Biol. 2009, 645, 301–306. [Google Scholar] [PubMed]
- Fontana, L.; Partridge, L.; Longo, V.D. Extending healthy life span-from yeast to humans. Science 2010, 328, 321–326. [Google Scholar] [CrossRef]
- Bauer, M.; Hamm, A.C.; Bonaus, M.; Jacob, A.; Jaekel, J.; Schorle, H.; Pankratz, M.J.; Katzenberger, J.D. Starvation response in mouse liver shows strong correlation with life-span-prolonging processes. Physiol. Genom. 2004, 17, 230–244. [Google Scholar] [CrossRef] [Green Version]
- Schweigert, F.J. Nutritional proteomics: Methods and concepts for research in nutritional science. Ann. Nutr. Metabol. 2007, 51, 99–107. [Google Scholar] [CrossRef]
- Bouwman, F.G.; Roos, B.; Rubio-Aliaga, I.; Crosley, L.K.; Duthie, S.J.; Mayer, C.; Horgan, G.; Polley, A.C.; Heim, C.; Coort, S.L.M.; et al. 2D-electrophoresis and multiplex immunoassay proteomic analysis of different body fluids and cellular components reveal known and novel markers for extended fasting. BMC Med. Genom. 2011, 4, 24. [Google Scholar] [CrossRef]
- Wang, H.; Ye, J. Regulation of energy balance by inflammation: Common theme in physiology and pathology. Rev. Endocr. Metabol. Disord. 2015, 16, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Schilling, B.; Rardin, M.J.; MacLean, B.X.; Zawadzka, A.M.; Frewen, B.E.; Cusack, M.P.; Sorensen, D.J.; Bereman, M.S.; Jing, E.; Wu, C.C.; et al. Gibson Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: Application to protein acetylation and phosphorylation. Mol. Cell. Proteom. 2012, 11, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.; Chang, C.Y.; Clough, T.; Broudy, D.; Killeen, T.; MacLean, B.; Vitek, O. MSstats: An R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 2014, 30, 2524–2526. [Google Scholar] [CrossRef] [PubMed]
- Jonckheere, A.I.; Renkema, G.H.; Bras, M.; Heuvel, L.P.; Hoischen, A.; Gilissen, C.; Nabuurs, S.B.; Huynen, M.A.; de Vries, M.C.; Smeitink, J.A.M.; et al. A complex V ATP5A1 defect causes fatal neonatal mitochondrial encephalopathy. Brain 2013, 136, 1544–1554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Formentini, L.; Pereira, M.P.; Sánchez-Cenizo, L.; Santacatterina, F.; Lucas, J.J.; Navarro, C.; Martínez-Serrano, A.; Cuezva, J.M. In vivo inhibition of the mitochondrial H+-ATP synthase in neurons promotes metabolic preconditioning. EMBO J. 2014, 33, 762–778. [Google Scholar] [CrossRef]
- Gusdon, A.M.; Fernandez-Bueno, G.A.; Wohlgemuth, S.; Fernández, J.; Chen, J.; Mathews, C.E. Respiration and substrate transport rates as well as reactive oxygen species production distinguish mitochondria from brain and liver. BMC Biochem. 2015, 16, 22. [Google Scholar] [CrossRef]
- Dröse, S.; Stepanova, A.; Galkin, A. Ischemic A/D transition of mitochondrial complex I and its role in ROS generation. Biochim. Biophys. Acta Bioenerg. 2016, 1857, 946–957. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Gómez, S.A.; Slamovits, C.H.; Dacks, J.B.; Wideman, J.G. The evolution of MICOS: Ancestral and derived functions and interactions. Commun. Integr. Biol. 2015, 8, e1094593. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.F.; Sun, L.H.; Zhang, R.; Zhang, Y.; Luo, Y.X.; Zheng, W.; Zhang, Z.Q.; Chen, H.Z.; Liu, D.P. Suppression of Mic60 compromises mitochondrial transcription and oxidative phosphorylation. Nat. Sci. Rep. 2015, 5, 7990. [Google Scholar] [CrossRef] [Green Version]
- Hara, M.R.; Cascio, M.B.; Sawa, A. GAPDH as a sensor of NO stress. Biochim. Biophys. Acta 2006, 1762, 502–509. [Google Scholar] [CrossRef] [Green Version]
- Salganik, M.; Sergeyev, V.G.; Shinde, V.; Meyers, C.A.; Gorbatyuk, M.S.; Lin, J.H.; Zolotukhin, S.; Gorbatyuk, O.S. The loss of glucose-regulated protein 78 (GRP78) during normal aging or from siRNA knockdown augments human alpha-synuclein (α-syn) toxicity to rat nigral neurons. Neurobiol. Aging 2015, 36, 2213–2223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuang, X.L.; Liu, F.; Chen, H.; Li, Y.; Liu, Y.; Xiao, J.; Shan, G.; Li, M.; Snider, B.J.; Qu, J.; et al. Reductions of the components of the calreticulin/calnexin quality-control system by proteasome inhibitors and their relevance in a rodent model of Parkinson’s disease. J. Neurosci. Res. 2014, 92, 1319–1329. [Google Scholar] [CrossRef]
- Lin, Q.; Cao, Y.; Gao, J. Serum calreticulin is a negative biomarker in patients with Alzheimer’s disease. Int. J. Mol. Sci. 2014, 15, 21740–21753. [Google Scholar] [CrossRef]
- Orbán-Németh, Z.; Simader, H.; Badurek, S.; Tranciková, A.; Propst, F. Microtubule-associated protein 1S, a short and ubiquitously expressed member of the microtubule-associated protein 1 family. J. Biol. Chem. 2005, 280, 2257–2265. [Google Scholar] [CrossRef] [PubMed]
- Ravin, R.; Blank, P.S.; Busse, B.; Ravin, N.; Vira, S.; Bezrukov, L.; Waters, H.; Guerrero-Cazares, H.; Quinones-Hinojosa, A.; Lee, P.R.; et al. Blast shockwaves propagate Ca2+ activity via purinergic astrocyte networks in human central nervous system cells. Nat. Sci. Rep. 2016, 6, 25713. [Google Scholar] [CrossRef]
- Greer, E.L.; Brunet, A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell 2009, 8, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Jiang, T.; Yu, Y.; Tang, H.; Lu, S.; Peng, Z.; Fan, J. Secernin-1 contributes to colon cancer progression through enhancing matrix metalloproteinase-2/9 exocytosis. Dis. Markers 2015, 2015, 230703. [Google Scholar] [CrossRef]
- James, S.R.; Cedeno, C.D.; Sharma, A.; Zhang, W.; Mohler, J.L.; Odunsi, K.; Wilson, E.M.; Karpf, A.R. DNA methylation and nucleosome occupancy regulate the cancer germline antigen gene MAGEA11. Epigenetics 2013, 8, 849–863. [Google Scholar] [CrossRef] [Green Version]
- Kubo, T.; Nakajima, H.; Nakatsuji, M.; Itakura, M.; Kaneshige, A.; Azuma, Y.T.; Inui, T.; Takeuchi, T. Active site cysteine-null glyceraldehyde-3-phosphate dehydrogenase (GAPDH) rescues nitric oxide-induced cell death. Nitric Oxide 2016, 53, 13–21. [Google Scholar] [CrossRef] [Green Version]
- Hamdan, F.F.; Saitsu, H.; Nishiyama, K.; Gauthier, J.; Dobrzeniecka, S.; Spiegelman, D.; Lacaille, J.C.; Décarie, J.C.; Matsumoto, N.; Rouleau, G.A.; et al. Identification of a novel in-frame de novo mutation in SPTAN1 in intellectual disability and pontocerebellar atrophy. Eur. J. Hum. Genet. 2012, 20, 796–800. [Google Scholar] [CrossRef] [Green Version]
- Tegha-Dunghu, J.; Bausch, E.; Neumann, B.; Wuensche, A.; Walter, T.; Ellenberg, J.; Gruss, O.J. MAP1S controls microtubule stability throughout the cell cycle in human cells. J. Cell Sci. 2014, 157, 5007–5013. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Shi, Q.; Zheng, S.; Luo, L.; Yuan, S.; Wang, X.; Cheng, Z.; Zhang, W. Role of α II spectrin breakdown products in the prediction of the severity and clinical outcome of acute traumatic brain injury. Exp. Ther. Med. 2016, 11, 2049–2053. [Google Scholar] [CrossRef] [PubMed]
- Ruzza, P.; Vitale, R.M.; Hussain, R.; Biondi, B.; Amodeo, P.; Sechi, G.; Siligardi, G. Interactions of GFAP with ceftriaxone and phenytoin: SRCD and molecular docking and dynamic simulation. Biochim. Biophys. Acta 2016, 1860, 2239–2248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zutphen, T.; Ciapaite, J.; Bloks, V.W.; Ackereley, C.; Gerding, A.; Jurdzinski, A.; de Moraes, R.A.; Zhang, L.; Wolters, J.C.; Bischoff, R.; et al. Malnutrition-associated liver steatosis and ATP depletion is caused by peroxisomal and mitochondrial dysfunction. J. Hepatol. 2016, 65, 1198–1208. [Google Scholar] [CrossRef] [Green Version]
- Beharry, C.; Cohen, L.S.; Di, J.; Ibrahim, K.; Briffa-Mirabella, S.; Alonso, A.D. Tau-induced neurodegeneration: Mechanisms and targets. Neurosci. Bull. 2014, 30, 346–358. [Google Scholar] [CrossRef]
- Larsen, P.L.; Clarke, C.F. Extension of life-span in Caenorhabditis elegans by diet lacking coenzyme. Q. Sci. Aging Knowl. Environ. 2002, 295, 120. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Hu, X.; Wei, Z.; Tam, K.Y. Cytoskeleton molecular motors: Structures and their functions in neuron. Int. J. Biol. Sci. 2016, 12, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Pirke, K.M.; Broocks, A.; Wilckens, T.; Marquard, R.; Schweiger, U. Starvation-induced hyperactivity in the rat: The role of endocrine and neurotransmitter changes. Neurosci. Biobehav. Rev. 1993, 17, 287–294. [Google Scholar] [CrossRef]
- Neely, A.N.; Mortimore, G.E. Localization of products of endogenous proteolysis in lysosomes of perfused rat liver. Biochem. Biophys. Res. Commun. 1974, 59, 680–687. [Google Scholar] [CrossRef]
- Seglen, P.O.; Gordon, P.B. 3-Methyladenine: Specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl. Acad. Sci. USA 1982, 79, 1889–1892. [Google Scholar] [CrossRef] [Green Version]
- Mejlvang, J.; Olsvik, H.; Svenning, S.; Bruun, J.A.; Abudu, Y.P.; Larsen, K.B.; Brech, A.; Hansen, T.E.; Brenne, H.; Hansen, T.; et al. Starvation induces rapid degradation of selective autophagy receptors by endosomal microautophagy. J. Cell. Biol. 2018, 217, 3640–3655. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, Y.; Kirisako, T.; Takao, T.; Satomi, Y.; Shimonishi, Y.; Ishihara, N.; Mizushima, N.; Tanida, I.; Kominami, E.; Ohsumi, M.; et al. A ubiquitin-like system mediates protein lipidation. Nature 2000, 408, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Hickey, R.W.; Bayir, H.; Watkins, S.C.; Tyurin, V.A.; Guo, F.; Kochanek, P.M.; Jenkins, L.W.; Ren, J.; Gibson, G.; et al. Starving neurons show sex differences in autophagy. J. Biol. Chem. 2009, 284, 2283–2396. [Google Scholar] [CrossRef] [PubMed]
- Hung, S.-Y.; Huang, W.-P.; Liou, H.-C.; Fu, W.-M. Autophagy protects neuron from Aβ-induced cytotoxicity. Autophagy 2009, 5, 502–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| BIOLOGICAL PROCESSES | |
| Cytoskeleton organization | CALR, GAPDH, GFAP, MAP1S, SPTAN1 |
| Organelle organization | CALR, GAPDH, GFAP, MAP1S, NDUFS1, SPTAN1 |
| MOLECULAR FUNCTIONS | |
| Proteins binding complex | GAPDH, GFAP, HSPA5, SPTAN1, UQCRC1 |
| Transport of protons for ATP synthase activity, rotational mechanisms | ATP5A1, ATP5B |
| Binding to unfolded proteins | CALR, HSPA5 |
| CELLULAR COMPONENTS | |
| Part of intracellular organelles | ATP5A1, ATP5B, CALR, GFAP, HSPA5, IMMT, MAP1S, NDUFS1, SCRN1, SPTAN1, UQCRC1 |
| Protein complex of inner mitochondrial membrane | ATP5A1, ATP5B, NDUFS1, UQCRC1 |
| Protein complex | ATP5A1, ATP5B, GAPDH, GFAP, HSPA5, MAP1S, NDUFS1, SPTAN1, UQCRC1 |
| Inner mitochondrial membrane | ATP5A1, ATP5B, IMMT, NDUFS1, UQCRC1 |
| Organelle membranes | ATP5A1, ATP5B, CALR, HSPA5, IMMT, NDUFS1, SCRN1,UQCRC1 |
| KEGG PATHWAYS | |
| Oxidative phosphorylation | ATP5A1, ATP5B, NDUFS1, UQCRC1 |
| Parkinson’s disease | ATP5A1, ATP5B, NDUFS1, UQCRC1 |
| Alzheimer’s disease | ATP5A1, ATP5B, NDUFS1, UQCRC1 |
| Huntington’s disease | ATP5A1, ATP5B, NDUFS1, UQCRC1 |
| Processing and introduction of antigens | CALR, HSPA5 |
 ) is expressed against control value (
) is expressed against control value (  ) that is considered with the value of 1. In all cases the differences between control and starvation were significant.
) that is considered with the value of 1. In all cases the differences between control and starvation were significant.
 ) is expressed against control value (
) is expressed against control value (  ) that is considered with the value of 1. In all cases the differences between control and starvation were significant.
) that is considered with the value of 1. In all cases the differences between control and starvation were significant.
| Spot a) | Homology | Species | Mw (kDa) | pI | # Peptides b) | Access Number | MOWSE Score c) | Coverage (%) d) |
|---|---|---|---|---|---|---|---|---|
| 501 | SCRN1 | Rattus norvegicus | 46.993 | 4.73 | 8 | sp|Q6AY84|SCRN1 | 68 | 21 |
| 701 | CALR | Rattus norvegicus | 48.136 | 4.37 | 16 | sp|P18418|CALR_ | 304 | 39 |
| 1402 | ATP5B | Oryctolagus cuniculus | 45.549 | 5.21 | 22 | tr|Q0QEN9|Q0QEN | 885 | 63 |
| 1404 | GFAP | Rattus norvegicus | 49.983 | 5.35 | 12 | sp|P47819|GFAP_ | 199 | 32 |
| 2708 | HSPA5 | Mus musculus | 72.492 | 5.01 | 34 | tr|Q9DC41|Q9DC4 | 646 | 53 |
| 2807 | SPTAN1 | Mus musculus | 98.016 | 5.17 | 26 | tr|Q3URW8|Q3URW | 210 | 32 |
| 2808 | IMMT | Rattus norvegicus | 86.689 | 5.62 | 13 | tr|A0A0G2JVH4|A | 75 | 18 |
| 3404 | UQCRC1 | Rattus norvegicus | 53.499 | 5.57 | 20 | sp|Q68FY0|QCR1_ | 327 | 39 |
| 3815 | NDUFS1 | Rattus norvegicus | 80.330 | 5.65 | 30 | sp|Q66HF1|NDUS1 | 386 | 46 |
| 4305 | MAP1S | Monodelphis domestica | 124.777 | 5.97 | 19 | tr|F7BK59|F7BK5 | 68 | 12 |
| 7506 | ATP5A1 | Rattus norvegicus | 59.889 | 9.29 | 11 | tr|F1LP05|F1LP0 | 92 | 23 |
| 7604 | MAGEA11 | Heterocephalus glaber | 55.034 | 5.27 | 13 | tr|G5C258|G5C25 | 66 | 19 |
| 8317 | GAPDH | Mus musculus | 36.072 | 8.44 | 13 | sp|P16858|G3P_M | 174 | 42 |
| 8319 | GAPDH | Mus musculus | 36.072 | 8.44 | 16 | sp|P16858|G3P_M | 283 | 50 |
| Abbreviation | Complete name | Functions |
|---|---|---|
| SCRN1 | Secernine-1 | Regulates exocytosis in mastocytes |
| CALR | Calreticuline | Calcium binding chaperone that promotes folding, oligomeric assembling and quality control in the endoplasmic reticulum by the calreticuline/calnexine cycle |
| ATP5B | β subunit of ATP synthase, mitochondrial precursor | ATP synthase located in the mitochondrial membrane that produce ATP from ADP |
| GFAP | Glial fibrillar acid protein | Specific cell target that, during development of central nervous system, distinguished astrocytes from other glia cells |
| HSPA5 | Heat-shock protein family A (Hsp70) member 5 | Facilitates the assembly of multimeric protein complex in the endoplasmic reticulum |
| SPTAN1 | Chain α of non-erythrocyte 1 spectrine | Interacts with calmodulin in a calcium-dependent manner and could participate in the calcium-dependent movement of cytoskeleton to membrane |
| IMMT | Mic60 subunit of mitochondrial contact site and cristae organizing system MICOS complex, protein of the inner mitochondrial membrane | Maintenance of architecture of the inner mitochondrial membrane and formation of contact sites with external membrane |
| UQCRC1 | Subunit 1 of mitochondrial cytochrome b-c1 | Component of the ubiquinole-cytochrome c reductase |
| NDUFS1 | 75kDa subunit of NADH-mitochondrial ubiquinone oxydoreductase | Core subunit of NADH dehydrogenase |
| MAP1S | Protein 1S associated to microtubule | Participate in the aggregation of mitochondria from cell death and the genomic breakdown |
| ATP5A1 | ATP synthase α subunit, mitochondrial precursor | ATP synthase from mitochondrial membrane |
| MAGEA11 | Antigen 11 associated to melanome | Co-regulator of androgen receptor that increases its activity. Involved in calcium homeostasis in endoplasmic reticulum |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | Key enzyme of glycolysis |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuevas-Fernández, B.; Fuentes-Almagro, C.; Peragón, J. Proteomics Analysis Reveals the Implications of Cytoskeleton and Mitochondria in the Response of the Rat Brain to Starvation. Nutrients 2019, 11, 219. https://doi.org/10.3390/nu11020219
Cuevas-Fernández B, Fuentes-Almagro C, Peragón J. Proteomics Analysis Reveals the Implications of Cytoskeleton and Mitochondria in the Response of the Rat Brain to Starvation. Nutrients. 2019; 11(2):219. https://doi.org/10.3390/nu11020219
Chicago/Turabian StyleCuevas-Fernández, Beatriz, Carlos Fuentes-Almagro, and Juan Peragón. 2019. "Proteomics Analysis Reveals the Implications of Cytoskeleton and Mitochondria in the Response of the Rat Brain to Starvation" Nutrients 11, no. 2: 219. https://doi.org/10.3390/nu11020219