Retrieving Forest Inventory Variables with Terrestrial Laser Scanning (TLS) in Urban Heterogeneous Forest
Abstract
:1. Introduction
2. Materials and Methods
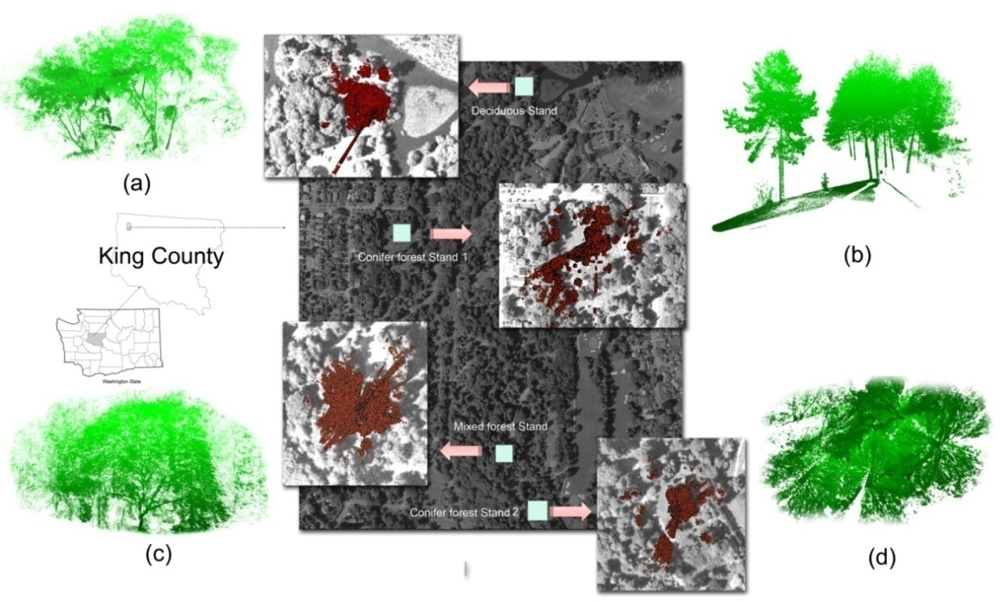
2.1. Study Area
2.2. Data Collection
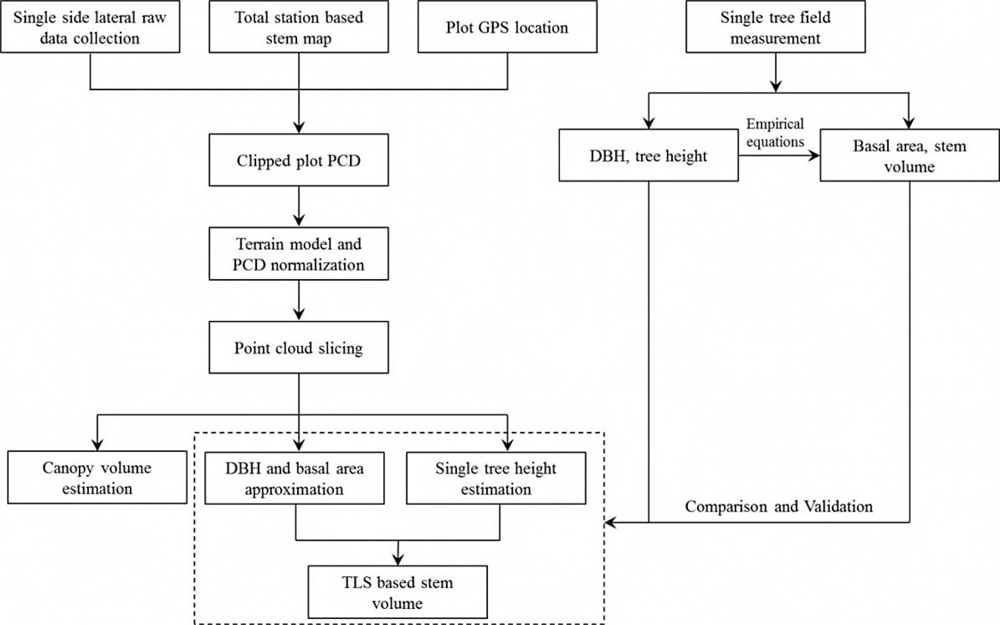
2.3. Algorithm Development
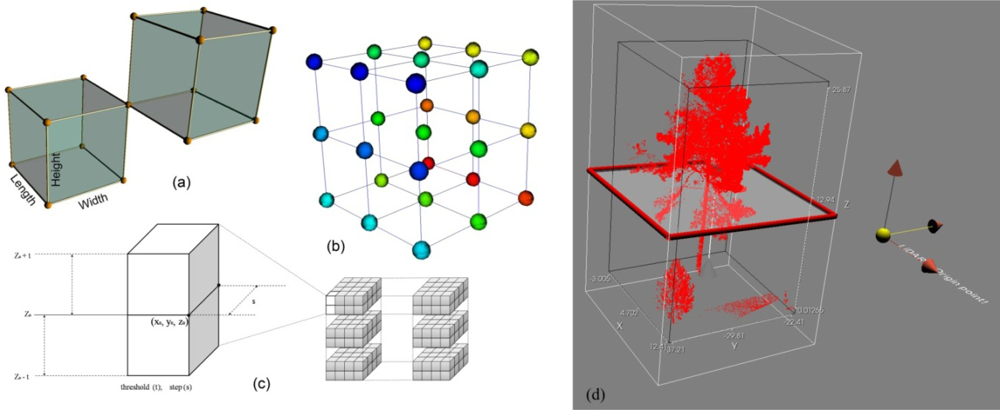
2.3.1. Voxel Modeling
2.3.2. Terrain Model for Normalizing PCD
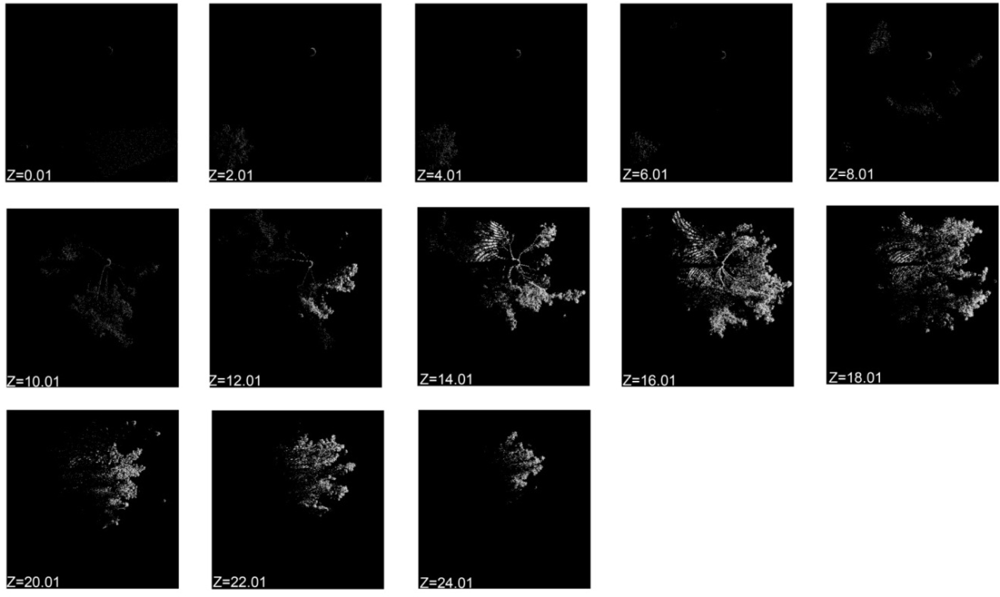
2.4. Point Cloud Slicing (PCS)
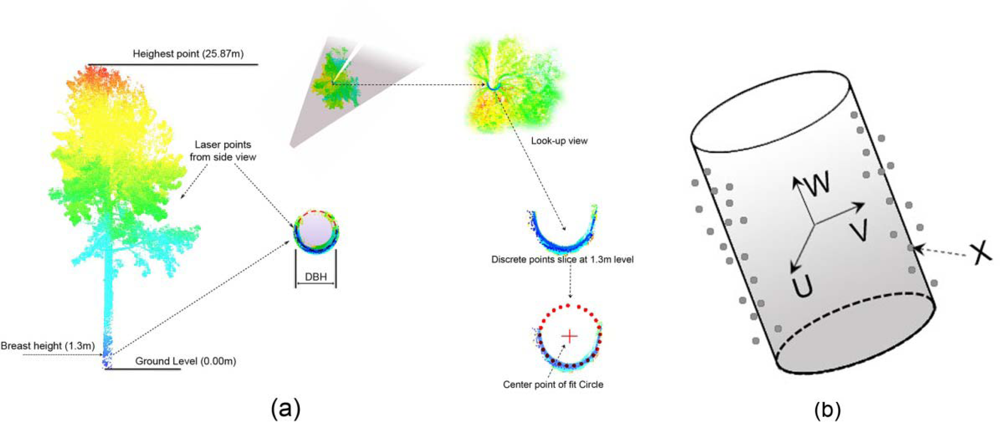
2.4.1. DBH, Basal Area, and Tree Heights Estimation
2.4.2. Stem and Canopy Volume Estimation
3. Results
3.1. Horizontal PCS for an Individual Tree
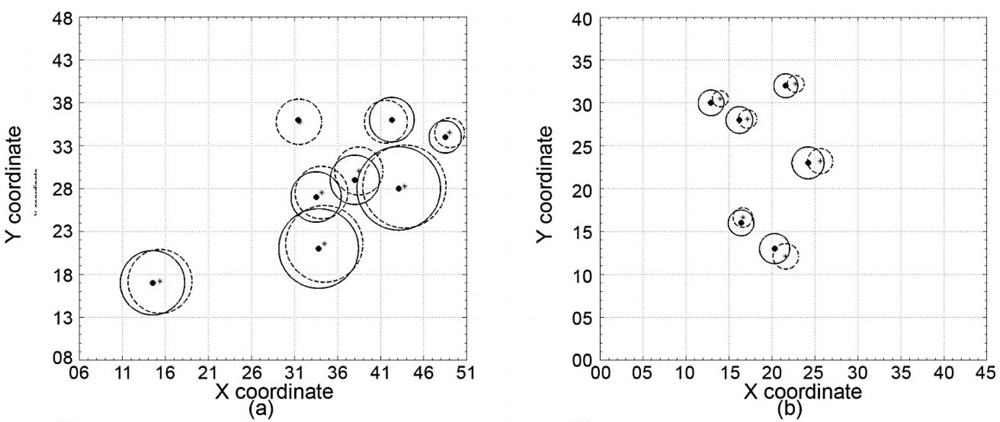
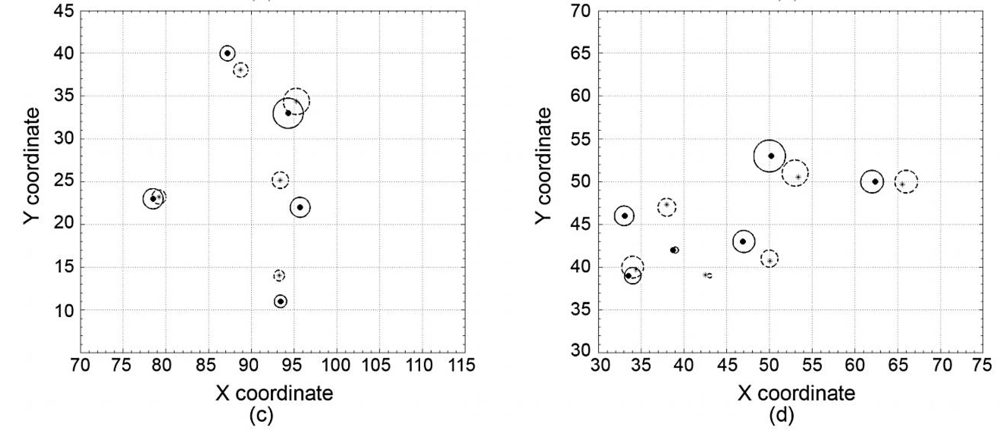
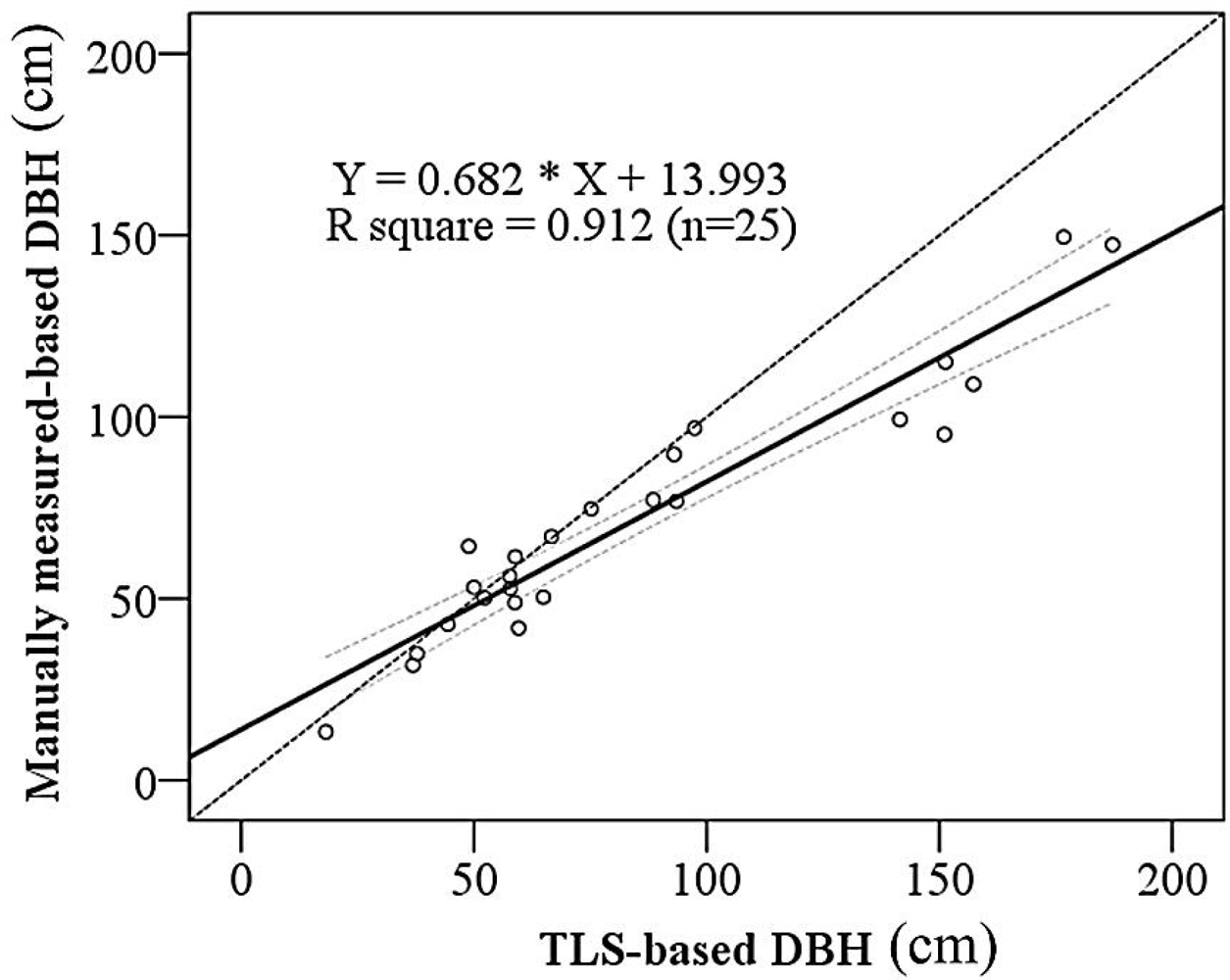
3.2. DBH and Stem Location Estimation
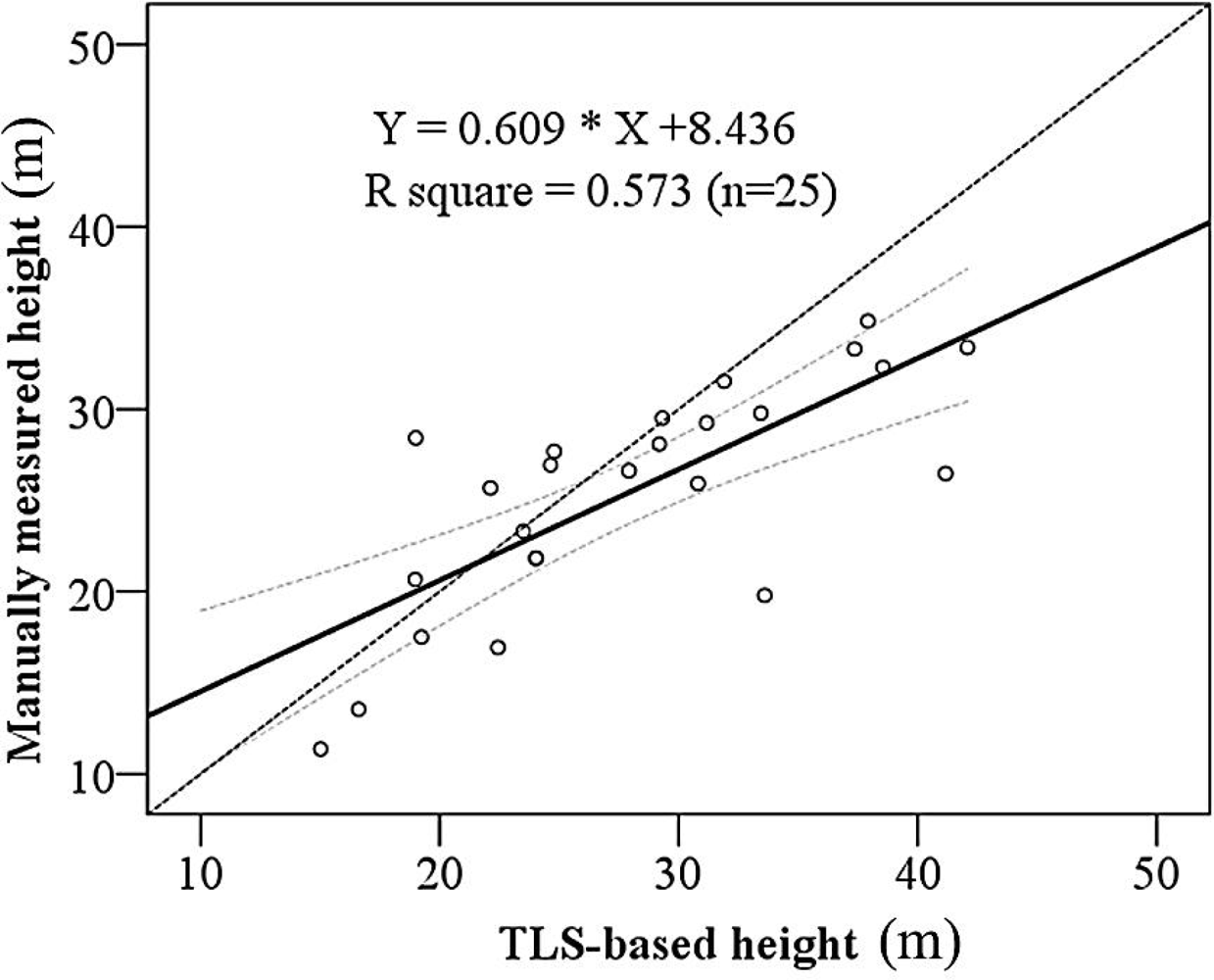
3.3. Basal Area and Tree Height Estimation
3.4. Canopy and Stem Volume Estimation
4. Discussion
5. Conclusions
Acknowledgments
References
- Fernandes, P.M. Combining forest structure data and fuel modelling to classify fire hazard in Portugal. Ann. For. Sci 2009, 66. [Google Scholar] [CrossRef]
- Zheng, G.; Chen, J.M.; Tian, Q.; Ju, W.M.; Xia, X.Q. Combining remote sensing imagery and forest age inventory for biomass mapping. J. Environ. Manage 2007, 85, 616–623. [Google Scholar]
- Wang, B.; Huang, J.Y.; Yang, X.S.; Zhang, B.A.; Liu, M.C. Estimation of biomass, net primary production and net ecosystem production of China’s forests based on the 1999–2003 national forest inventory. Scand. J. For. Res 2010, 25, 544–553. [Google Scholar]
- Van Deusen, P. Carbon sequestration potential of forest land: Management for products and bioenergy versus preservation. Biomass Bioenergy 2010, 34, 1687–1694. [Google Scholar]
- Thony, P.; Mabee, W.; Kozak, R.; Bull, G. A characterization of the british columbia log home and timber frame manufacturing sector. Forest. Chron 2006, 82, 77–83. [Google Scholar]
- Cottone, N.; Ettl, G.J. Estimating populations of whitebark pine in mount rainier national park, washington, using aerial photography. Northwest Sci 2001, 75, 397–406. [Google Scholar]
- Barclay, H.J.; Li, C.; Benson, L.; Taylor, S.; Shore, T. Effects of fire return rates on traversability of lodgepole pine forests for mountain pine beetle (coleoptera:Scolytidae) and the use of patch metrics to estimate traversability. Can. Entomol 2005, 137, 566–583. [Google Scholar]
- Pothier, D. Twenty-year results of precommercial thinning in a balsam fir stand. Forest Ecol. Manage 2002, 168, 177–186. [Google Scholar]
- Patenaude, G.; Milne, R.; Dawson, T.P. Synthesis of remote sensing approaches for forest carbon estimation: Reporting to the Kyoto Protocol. Environ. Sci. Policy 2005, 8, 161–178. [Google Scholar]
- Kim, Y.; Yang, Z.Q.; Cohen, W.B.; Pflugmacher, D.; Lauver, C.L.; Vankat, J.L. Distinguishing between live and dead standing tree biomass on the north rim of Grand Canyon National Park, USA using small-footprint lidar data. Remote Sens. Environ 2009, 113, 2499–2510. [Google Scholar]
- Chen, J.M.; Chen, X.Y.; Ju, W.M.; Geng, X.Y. Distributed hydrological model for mapping evapotranspiration using remote sensing inputs. J. Hydrol 2005, 305, 15–39. [Google Scholar]
- Nikolov, N.; Zeller, K.F. Modeling coupled interactions of carbon, water, and ozone exchange between terrestrial ecosystems and the atmosphere. I: Model description. Environ. Pollut 2003, 124, 231–246. [Google Scholar]
- Richardson, J.J.; Moskal, L.M.; Kim, S.H. Modeling approaches to estimate effective leaf area index from aerial discrete-return lidar. Agric. Forest Meteorol 2009, 149, 1152–1160. [Google Scholar]
- Moskal, L.M.; Styers, D.M.; Halabisky, M. Monitoring urban tree cover using object-based image analysis and public domain remotely sensed data. Remote Sens 2011, 3, 2243–2262. [Google Scholar]
- Henning, J.G.; Radtke, P.J. Ground-based laser imaging for assessing three-dimensional forest canopy structure. Photogramm. Eng. Remote Sensing 2006, 72, 1349–1358. [Google Scholar]
- Niinemets, U. Photosynthesis and resource distribution through plant canopies. Plant Cell Environ 2007, 30, 1052–1071. [Google Scholar]
- Utsugi, H.; Araki, M.; Kawasaki, T.; Ishizuka, M. Vertical distributions of leaf area and inclination angle, and their relationship in a 46-year-old chamaecyparis obtusa stand. Forest Ecol. Manage 2006, 225, 104–112. [Google Scholar]
- Li, F.S.; Cohen, S.; Naor, A.; Kang, S.Z.; Erez, A. Studies of canopy structure and water use of apple trees on three rootstocks. Agric. Water Manage 2002, 55, 1–14. [Google Scholar]
- Macfarlane, C.; Grigg, A.; Evangelista, C. Estimating forest leaf area using cover and fullframe fisheye photography: Thinking inside the circle. Agric. Forest Meteorol 2007, 146, 1–12. [Google Scholar]
- Zhang, S.Y.; Lei, Y.C.; Bowling, C. Quantifying stem quality characteristics in relation to initial spacing and modeling their relationship with tree characteristics in black spruce (picea mariana). Northern J. Appl. Forest 2005, 22, 85–93. [Google Scholar]
- Evans, D.L.; Roberts, S.D.; Parker, R.C. Lidar—A new tool for forest measurements? Forest Chron 2006, 82, 211–218. [Google Scholar]
- van Leeuwen, M.; Nieuwenhuis, M. Retrieval of forest structural parameters using lidar remote sensing. Eur. J. For. Res 2010, 129, 749–770. [Google Scholar]
- Jang, J.D.; Payan, V.; Viau, A.A.; Devost, A. The use of airborne lidar for orchard tree inventory. Int. J. Remote Sens 2008, 29, 1767–1780. [Google Scholar]
- Naesset, E. Airborne laser scanning as a method in operational forest inventory: Status of accuracy assessments accomplished in Scandinavia. Scand. J. Forest Res 2007, 22, 433–442. [Google Scholar]
- Maas, H.G.; Bienert, A.; Scheller, S.; Keane, E. Automatic forest inventory parameter determination from terrestrial laser scanner data. Int. J. Remote Sens 2008, 29, 1579–1593. [Google Scholar]
- Erdody, T.L.; Moskal, L.M. Fusion of lidar and imagery for estimating forest canopy fuels. Remote Sens. Environ 2010, 114, 725–737. [Google Scholar]
- Anderson, J.E.; Plourde, L.C.; Martin, M.E.; Braswell, B.H.; Smith, M.L.; Dubayah, R.O.; Hofton, M.A.; Blair, J.B. Integrating waveform lidar with hyperspectral imagery for inventory of a northern temperate forest. Remote Sens. Environ 2008, 112, 1856–1870. [Google Scholar]
- Hyyppa, J.; Hyyppa, H.; Leckie, D.; Gougeon, F.; Yu, X.; Maltamo, M. Review of methods of small-footprint airborne laser scanning for extracting forest inventory data in boreal forests. Int. J. Remote Sens 2008, 29, 1339–1366. [Google Scholar]
- Hill, R.A.; Thomson, A.G. Mapping woodland species composition and structure using airborne spectral and lidar data. Int. J. Remote Sens 2005, 26, 3763–3779. [Google Scholar]
- Vaughn, N.R.; Moskal, L.M.; Turnblom, E.C. Fourier transformation of waveform lidar for species recognition. Remote Sens. Lett 2011, 2, 347–356. [Google Scholar]
- Watt, P.J.; Donoghue, D.N.M. Measuring forest structure with terrestrial laser scanning. Int. J. Remote Sens 2005, 26, 1437–1446. [Google Scholar]
- Tansey, K.; Selmes, N.; Anstee, A.; Tate, N.J.; Denniss, A. Estimating tree and stand variables in a corsican pine woodland from terrestrial laser scanner data. Int. J. Remote Sens 2009, 30, 5195–5209. [Google Scholar]
- Lovell, J.L.; Jupp, D.L.B.; Newnham, G.J.; Culvenor, D.S. Measuring tree stem diameters using intensity profiles from ground-based scanning lidar from a fixed viewpoint. ISPRS J. Photogramm 2011, 66, 46–55. [Google Scholar]
- Huang, H.B.; Li, Z.; Gong, P.; Cheng, X.A.; Clinton, N.; Cao, C.X.; Ni, W.J.; Wang, L. Automated methods for measuring DBH and tree heights with a commercial scanning lidar. Photogramm. Eng. Remote Sensing 2011, 77, 219–227. [Google Scholar]
- Murphy, G. Determining stand value and log product yields using terrestrial lidar and optimal bucking: A case study. J. Forest 2008, 106, 317–324. [Google Scholar]
- Omasa, K.; Urano, Y.; Oguma, H.; Fujinuma, Y. Mapping of tree position of larix leptolepis woods and estimation of diameter at breast height (DBH) and biomass of the trees using range data measured by a portable scanning lidar. J. Remote Sens. Soc. Jpn 2002, 22, 550–557. [Google Scholar]
- Hopkinson, C.; Chasmer, L. Testing lidar models of fractional cover across multiple forest ecozones. Remote Sens. Environ 2009, 113, 275–288. [Google Scholar]
- Weiss, J. Application and statistical analysis of terrestrial laser scanning and forest growth simulations to determine selected characteristics of douglas-fir stands. Felia Forestalia Polonica 2009, 51, 123–137. [Google Scholar]
- Yao, T.; Yang, X.; Zhao, F.; Wang, Z.; Zhang, Q.; Jupp, D.; Lovell, J.; Culvenor, D.; Newnham, G.; Ni-Meister, W.; et al. Measuring forest structure and biomass in New England forest stands using echidna ground-based lidar. Remote Sens. Environ 2011, 115, 2965–2974. [Google Scholar]
- Popescu, S.C. Estimating biomass of individual pine trees using airborne lidar. Biomass Bioenergy 2007, 31, 646–655. [Google Scholar]
- Jupp, D.L.B.; Culvenor, D.S.; Lovell, J.L.; Newnham, G.J.; Strahler, A.H.; Woodcock, C.E. Estimating forest lai profiles and structural parameters using a ground-based laser called “echidna (r)”. Tree Physiol 2009, 29, 171–181. [Google Scholar]
- Kato, A.; Moskal, L.M.; Schiess, P.; Swanson, M.E.; Calhoun, D.; Stuetzle, W. Capturing tree crown formation through implicit surface reconstruction using airborne lidar data. Remote Sens. Environ 2009, 113, 1148–1162. [Google Scholar]
- van Aardt, J.A.N.; Wynne, R.H.; Scrivani, J.A. Lidar-based mapping of forest volume and biomass by taxonomic group using structurally homogenous segments. Photogramm. Eng. Remote Sensing 2008, 74, 1033–1044. [Google Scholar]
- Zheng, G.; Moskal, L.M. Retrieving leaf area index (LAI) using remote sensing: Theories, methods and sensors. Sensors 2009, 9, 2719–2745. [Google Scholar]
- Chen, Q.; Gong, P.; Baldocchi, D.; Xie, G. Filtering airborne laser scanning data with morphological methods. Photogramm. Eng. Remote Sensing 2007, 73, 175–185. [Google Scholar]
- Kraus, K.; Pfeifer, N. Determination of terrain models in wooded areas with airborne laser scanner data. ISPRS J. Photogramm 1998, 53, 193–203. [Google Scholar]
- Kobler, A.; Pfeifer, N.; Ogrinc, P.; Todorovski, L.; Ostir, K.; Dzeroski, S. Repetitive interpolation: A robust algorithm for dtm generation from aerial laser scanner data in forested terrain. Remote Sens. Environ 2007, 108, 9–23. [Google Scholar]
- Thies, M.; Pfeifer, N.; Winterhalder, D.; Gorte, B.G.H. Three-dimensional reconstruction of stems for assessment of taper, sweep and lean based on laser scanning of standing trees. Scand. J. Forest Res 2004, 19, 571–581. [Google Scholar]
- Schneider, P.J.; Eberly, D.H. Appendix A: Numerical methods. In Geometric Tools for Computer Graphics; Morgan Kaufmann Publishers: Amsterdam, The Netherlands, 2003; pp. 882–889. [Google Scholar]
- Eberly, D. Fitting 3D Data with a Cylinder; Geometric Tools, LLC.: Scottsdale, AZ, USA, 2008. Available online: http://www.geometrictools.com/Documentation/CylinderFitting.pdf (accessed on 5 February 2011).
- Bouriaud, O.; Soudani, K.; Breda, N. Leaf area index from litter collection: Impact of specific leaf area variability within a beech stand. Can. J. Remote Sens 2003, 29, 371–380. [Google Scholar]
- Jung, S.-E.; Kwak, D.-A.; Park, T.; Lee, W.-K.; Yoo, S. Estimating crown variables of individual trees using airborne and terrestrial laser scanners. Remote Sens 2011, 3, 2346–2363. [Google Scholar]
- Lin, Y.; Jaakkola, A.; Hyyppä, J.; Kaartinen, H. From TLS to VLS: Biomass estimation at individual tree level. Remote Sens 2010, 2, 1864–1879. [Google Scholar]
- Van der Zande, D.; Hoet, W.; Jonckheere, I.; van Aardt, J.; Coppin, P. Influence of measurement set-up of ground-based lidar for derivation of tree structure. Agric. Forest. Meteorol 2006, 141, 147–160. [Google Scholar]
- Seielstad, C.; Stonesifer, C.; Rowell, E.; Queen, L. Deriving fuel mass by size class in Douglas-fir (Pseudotsuga menziesii) using terrestrial laser scanning. Remote Sens 2011, 3, 1691–1709. [Google Scholar]
- Pesci, A.; Teza, G.; Bonali, E. Terrestrial laser scanner resolution: Numerical simulations and experiments on spatial sampling optimization. Remote Sens 2011, 3, 167–184. [Google Scholar]

| Conifer-1 | Conifer-2 | Deciduous | Mixed | |||||
|---|---|---|---|---|---|---|---|---|
| Field-TBA | TLS-TBA | Field-TBA | TLS-TBA | Field-TBA | TLS-TBA | Field-TBA | TLS-TBA | |
| 0.1122 | 0.0951 | 1.7979 | 1.0405 | 0.279 | 0.1379 | 0.6866 | 0.4632 | |
| 0.2148 | 0.1979 | 2.4522 | 1.7554 | 0.1548 | 0.1445 | 0.3308 | 0.1995 | |
| — | 0.2215 | 2.7523 | 1.7064 | 0.6151 | 0.4681 | 0.2624 | 0.219 | |
| 0.2615 | 0.2481 | 1.9433 | 0.9331 | 0.2715 | 0.1878 | 0.3494 | 0.3536 | |
| 0.7451 | 0.7375 | 1.7932 | 0.7118 | 0.1069 | 0.0789 | 0.026 | 0.0139 | |
| 0.6793 | 0.6319 | 1.5725 | 0.7744 | 0.1878 | 0.3257 | |||
| 0.2725 | 0.2971 | |||||||
| 0.4441 | 0.4383 | |||||||
| SBAA | 2.7295 | 2.8673 | 12.3116 | 6.9217 | 1.4275 | 1.0172 | 1.8431 | 1.575 |
| Coefficient | Estimate | Standard Error | t-Value | P-Value | |
|---|---|---|---|---|---|
| DBH-prediction | Intercept | 13.993 | 4.299 | 3.255 | 0.003 |
| Manually measured-based DBH | 0.682 | 0.044 | 15.407 | 0.000 | |
| Height-prediction | Intercept | 8.436 | 3.177 | 2.656 | 0.014 |
| TLS-based tree height | 0.609 | 0.110 | 5.552 | 0.000 | |
| BP Value | Degree of Freedom | P-Value | Chi-Square Value (95% level) | |
|---|---|---|---|---|
| TLS-DBH prediction | 1.725 | 1 | 0.189 | 3.841 |
| TLS-height prediction | 0.293 | 1 | 0.585 | 3.841 |
| Conifer-1 | Conifer-2 | Deciduous | Mixed | |||||
|---|---|---|---|---|---|---|---|---|
| Field-STV | TLS-STV | Field-STV | TLS-STV | Field-STV | TLS-STV | Field--STV | TLS-STV | |
| 0.88 | 0.74 | 22.73 | 13.15 | 4.40 | 2.97 | 2.23 | 1.10 | |
| — | 1.69 | 26.08 | 18.67 | 3.22 | 1.94 | 0.78 | 0.72 | |
| 1.58 | 1.53 | 35.37 | 21.93 | 2.94 | 2.45 | 4.93 | 3.75 | |
| 2.43 | 2.20 | 24.20 | 11.62 | 3.42 | 3.46 | 2.03 | 1.40 | |
| 7.65 | 6.37 | 24.61 | 9.77 | 0.16 | 0.09 | 0.59 | 0.44 | |
| 7.06 | 6.16 | 22.06 | 10.86 | 1.55 | 2.69 | |||
| 3.04 | 2.95 | |||||||
| 3.65 | 3.94 | |||||||
| FPV t | 21.40 | 25.58+0.0045 | 155.06 | 86.01+0.0038 | 15.69 | 13.60+0.0017 | 10.56 | 7.42+0.0037 |
Share and Cite
Moskal, L.M.; Zheng, G. Retrieving Forest Inventory Variables with Terrestrial Laser Scanning (TLS) in Urban Heterogeneous Forest. Remote Sens. 2012, 4, 1-20. https://doi.org/10.3390/rs4010001
Moskal LM, Zheng G. Retrieving Forest Inventory Variables with Terrestrial Laser Scanning (TLS) in Urban Heterogeneous Forest. Remote Sensing. 2012; 4(1):1-20. https://doi.org/10.3390/rs4010001
Chicago/Turabian StyleMoskal, L. Monika, and Guang Zheng. 2012. "Retrieving Forest Inventory Variables with Terrestrial Laser Scanning (TLS) in Urban Heterogeneous Forest" Remote Sensing 4, no. 1: 1-20. https://doi.org/10.3390/rs4010001






