Multi-Species Individual Tree Segmentation and Identification Based on Improved Mask R-CNN and UAV Imagery in Mixed Forests
Abstract
:1. Introduction
2. Study Areas and Material
2.1. Study Site
2.2. Field Data
2.3. Individual Tree Crown Dataset
2.3.1. Orthophoto Map
2.3.2. Sample Labels
3. Methods
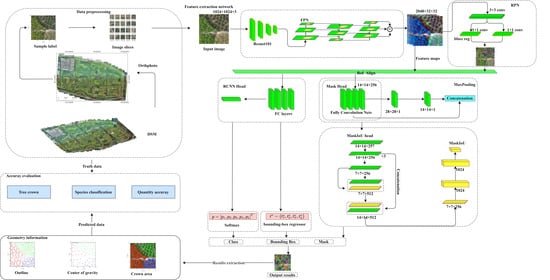
3.1. Overall Workflow
3.2. Mask R-CNN and the Improved Model
3.2.1. Mask R-CNN
3.2.2. Network Improvements
- (a)
- Modification of the fusion style
- (b)
- Evolution of loss function Lmask
3.3. Outline and Center Extraction
| if (fij = 1 & fi,j−1 = 0): |
| { |
| (i,j) is the starting point of the outer boundary; |
| (i2,j2) = (i,j−1); |
| } |
| else |
| { |
| Continue scanning grating; |
| } |
3.4. Evaluation Index
3.5. Network Training and a Comparison with Different Models
4. Results
4.1. Accuracy Evaluation of Individual Tree Crown Segmentation
4.2. Species Identification and Classification Accuracy Evaluation
4.3. Accuracy Evaluation of Tree Count Detection
5. Discussion
5.1. Comparison of the Segmentation and Detection Performance of Different Networks
5.2. Comparison of Training Time and Loss
5.3. The Offset Value in the Center of Gravity and the Bounding Box
5.4. Segmentation Results at Different Brightness Levels
5.5. False Segmentation
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wagner, F.H.; Ferreira, M.P.; Sanchez, A.; Hirye, M.C.M.; Zortea, M.; Gloor, E.; Phillips, O.L.; Filho, C.R.d.S.; Shimabukuro, Y.E.; Aragão, L.E.O.C. Individual Tree Crown Delineation in a Highly Diverse Tropical Forest Using Very High Resolution Satellite Images. ISPRS J. Photogramm. Remote Sens. 2018, 145, 362–377. [Google Scholar] [CrossRef]
- Lindquist, E.J.; D’Annunzio, R.; Gerrand, A.; MacDicken, K.; Achard, F.; Beuchle, R.; Brink, A.; Eva, H.D.; Mayaux, P.; San-Miguel-Ayanz, J.; et al. Global Forest Land-Use Change 1990–2005; FAO Forestry Paper: Rome, Italy, 2012. [Google Scholar]
- Crowther, T.W.; Glick, H.B.; Covey, K.R.; Bettigole, C.; Maynard, D.S.; Thomas, S.M.; Smith, J.R.; Hintler, G.; Duguid, M.C.; Amatulli, G.; et al. Mapping Tree Density at a Global Scale. Nature 2015, 525, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Diez, Y.; Kentsch, S.; Fukuda, M.; Caceres, M.L.L.; Moritake, K. Deep Learning in Forestry Using UAV-Acquired RGB Data: A Practical Review. Remote Sens. 2021, 13, 2837. [Google Scholar] [CrossRef]
- Wang, L.; Gong, P.; Biging, G.S. Individual Tree-Crown Delineation and Treetop Detection in High-Spatial-Resolution Aerial Imagery. Photogramm. Eng. Remote Sens. 2004, 70, 351–358. [Google Scholar] [CrossRef] [Green Version]
- Santos, A.A.d.; Marcato, J., Jr.; Araújo, M.S.; Di Martini, D.R.; Tetila, E.C.; Siqueira, H.L.; Aoki, C.; Eltner, A.; Matsubara, E.T.; Pistori, H.; et al. Assessment of CNN-Based Methods for Individual Tree Detection on Images Captured by RGB Cameras Attached to UAVs. Sensors 2019, 19, 3595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miraki, M.; Sohrabi, H.; Fatehi, P.; Kneubuehler, M. Individual Tree Crown Delineation from High-Resolution UAV Images in Broadleaf Forest. Ecol. Inform. 2021, 61, 101207. [Google Scholar] [CrossRef]
- Dainelli, R.; Toscano, P.; Gennaro, S.F.D.; Matese, A. Recent Advances in Unmanned Aerial Vehicle Forest Remote Sensing—A Systematic Review. Part I: A General Framework. Forests 2021, 12, 327. [Google Scholar] [CrossRef]
- Harikumar, A.; Bovolo, F.; Bruzzone, L. A Local Projection-Based Approach to Individual Tree Detection and 3-D Crown Delineation in Multistoried Coniferous Forests Using High-Density Airborne LiDAR Data. IEEE Trans. Geosci. Remote Sens. 2019, 57, 1168–1182. [Google Scholar] [CrossRef]
- Zhang, B.; Zhao, L.; Zhang, X. Three-Dimensional Convolutional Neural Network Model for Tree Species Classification Using Airborne Hyperspectral Images. Remote Sens. Environ. 2020, 247, 111938. [Google Scholar] [CrossRef]
- Modzelewska, A.; Kamińska, A.; Fassnacht, F.E.; Stereńczak, K. Multitemporal Hyperspectral Tree Species Classification in the Biaowiea Forest World Heritage Site. Forestry 2021, 94, 464–476. [Google Scholar] [CrossRef]
- Liu, K.; Wang, A.; Zhang, S.; Zhu, Z.; Bi, Y.; Wang, Y.; Du, X. Tree Species Diversity Mapping Using UAS-Based Digital Aerial Photogrammetry Point Clouds and Multispectral Imageries in a Subtropical Forest Invaded by Moso Bamboo (Phyllostachys Edulis). Int. J. Appl. Earth Obs. Geoinform. 2021, 104, 102587. [Google Scholar] [CrossRef]
- Tochon, G.; Féret, J.B.; Valero, S.; Martin, R.E.; Knapp, D.E.; Salembier, P.; Chanussot, J.; Asner, G.P. On the Use of Binary Partition Trees for the Tree Crown Segmentation of Tropical Rainforest Hyperspectral Images. Remote Sens. Env. 2015, 159, 318–331. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Cai, X.; Lellmann, J.; Dalponte, M.; Malhi, Y.; Butt, N.; Morecroft, M.; Schönlieb, C.-B.; Coomes, D.A. Individual Tree Species Classification from Airborne Multisensor Imagery Using Robust PCA. IEEE J. Sel. Top. Appl. Earth Obser. Remote Sens. 2016, 9, 2554–2567. [Google Scholar] [CrossRef] [Green Version]
- Jing, L.; Hu, B.; Noland, T.; Li, J. An Individual Tree Crown Delineation Method Based on Multi-Scale Segmentation of Imagery. ISPRS J. Photogramm. Remote Sens. 2012, 70, 88–98. [Google Scholar] [CrossRef]
- Zhang, J.; Sohn, G.; Bredif, M. A Hybrid Framework for Single Tree Detection from Airborne Laser Scanning Data: A Case Study in Temperate Mature Coniferous Forests in Ontario, Canada. J. Photogramm. Remote Sens. 2014, 98, 44–57. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Im, J.; Lindi, J.Q. A Novel Transferable Individual Tree Crown Delineation Model Based on Fishing Net Dragging and Boundary Classification. ISPRS J. Photogramm. Remote Sens. 2015, 110, 34–47. [Google Scholar] [CrossRef]
- Cho, M.A.; Malahlela, O.; Ramoelo, A. Assessing the Utility Worldview-2 Imagery for Tree Species Mapping in South African Subtropical Humid Forest and the Conservation Implications: Dukuduku Forest Patch as Case Study. Int. J. Appl. Earth Obser. Geoinform. 2015, 38, 349–357. [Google Scholar] [CrossRef]
- Mutanga, O.; Adam, E.; Cho, M.A. High Density Biomass Estimation for Wetland Vegetation Using Worldview-2 Imagery and Random Forest Regression Algorithm. Int. J. Appl. Earth Obser. Geoinform. 2012, 18, 399–406. [Google Scholar] [CrossRef]
- Schiefer, F.; Kattenborn, T.; Frick, A.; Frey, J.; Schall, P.; Koch, B.; Schmidtlein, S. Mapping Forest Tree Species in High Resolution UAV-Based RGB-Imagery by Means of Convolutional Neural Networks. ISPRS J. Photogramm. Remote Sens. 2020, 170, 205–215. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Knapp, D.E.; Tupayachi, R.; Anderson, C.B.; Sinca, F.; Vaughn, N.R.; Llactayo, W. Airborne Laser-Guided Imaging Spectroscopy to Mapforest Trait Diversity and Guide Conservation. Science 2017, 355, 385–389. [Google Scholar] [CrossRef]
- Su, A.; Qi, J.; Huang, H. Indirect Measurement of Forest Canopy Temperature by Handheld Thermal Infrared Imager through Upward Observation. Remote Sens. 2020, 12, 3559. [Google Scholar] [CrossRef]
- Brandt, M.; Tucker, C.J.; Kariryaa, A.; Ransmussen, K.; Abel, C.; Small, J.; Chave, J.; Rasmussen, L.V.; Hiernaux, P.; Diouf, A.A.; et al. An Unexpectedly Large Count of Trees in the West African Sahara and Sahel. Nature 2020, 587, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Chadwick, A.J.; Goodbody, T.R.H.; Coops, N.C.; Hervieux, A.; Bater, C.W.; Martens, L.A.; White, B.; Röeser, D. Automatic Delineation and Height Measurement of Regenerating Conifer Crowns under Leaf-Off Conditions Using UAV Imagery. Remote Sens. 2020, 12, 4104. [Google Scholar] [CrossRef]
- Fujimoto, A.; Haga, C.; Matsui, T.; Machimura, T.; Hayashi, K.; Sugita, S.; Takagi, H. An End to End Process Development for UAV-SfM Based Forest Monitoring: Individual Tree Detection, Species Classification and Carbon Dynamics Simulation. Forests 2019, 10, 680. [Google Scholar] [CrossRef] [Green Version]
- Egli, S.; Höpke, M. CNN-Based Tree Species Classification Using High Resolution RGB Image Data from Automated UAV Observations. Remote Sens. 2020, 12, 3892. [Google Scholar] [CrossRef]
- Tran, D.Q.; Park, M.; Jung, D.; Park, S. Damage-Map Estimation Using UAV Images and Deep Learning Algorithms for Disaster Management System. Remote Sens. 2020, 12, 4169. [Google Scholar] [CrossRef]
- Safonova, A.; Tabik, S.; Alcaraz-Segura, D.; Rubtsov, A.; Maglinets, Y.; Herrera, F. Detection of Fir Trees (Abies Sibirica) Damaged by the Bark Beetle in Unmanned Aerial Vehicle Images with Deep Learning. Remote Sens. 2019, 11, 643. [Google Scholar] [CrossRef] [Green Version]
- Cao, K.; Zhang, X. An Improved Res-UNet Model for Tree Species Classification Using Airborne High-Resolution Images. Remote Sens. 2020, 12, 1128. [Google Scholar] [CrossRef] [Green Version]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015. [Google Scholar] [CrossRef] [Green Version]
- Osco, L.P.; Arruda, M.S.; Gonçalves, D.N.; Dias, A.; Batistoti, J.; Souza, M.; Gomes, F.D.G.; Ramos, A.P.M.; Jorge, L.A.C.; Liesenberg, V.; et al. A CNN Approach to Simultaneously Count Plants and Detect Plantation-Rows from UAV Imagery. ISPRS J. Photogramm. Remote Sens. 2021, 174, 1–17. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-CNN. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 42, 386–397. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Yin, Q.; Zheng, X.; Chen, Z. Remote Sensing Image Building Detection Method Based on Mask R-CNN. Complex Intell. Syst. 2021, 1–9. [Google Scholar] [CrossRef]
- Zheng, J.; Fu, H.; Li, W.; Wu, W.; Yu, L.; Yuan, S.; Yuk, W.; William, T.; Pang, T.K.; Kanniah, K.D.; et al. Growing Status Observation for Oil Palm Trees Using Unmanned Aerial Vehicle (UAV) Images. J. Photogramm. Remote Sens. 2021, 173, 95–121. [Google Scholar] [CrossRef]
- Braga, J.R.G.; Peripato, V.; Dalagnol, R.; Ferreira, M.P.; Tarabalka, Y.; Aragão, L.E.O.C.; Velho, H.F.C.; Shiguemori, E.H.; Wagner, F.H. Tree Crown Delineation Algorithm Based on a Convolutional Neural Network. Remote Sens. 2020, 12, 1288. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; Wang, X.; Zhang, H.; Yin, Z.; Su, D.; Xu, J. A Mask R-CNN Based Particle Identification for Quantitative Shape Evaluation of Granular Materials. Powder Technol. 2021, 392, 296–305. [Google Scholar] [CrossRef]
- Safonova, A.; Guirado, E.; Maglinets, Y.; Domingo, A.-S.; Tabik, S. Olive Tree Biovolume from UAV Multi-Resolution Image Segmentation with Mask R-CNN. Sensors 2021, 21, 1617. [Google Scholar] [CrossRef]
- Cao, X.; Pan, J.S.; Wang, Z.; Sun, Z.; Haq, A.; Deng, W.; Yang, S. Application of Generated Mask Method Based on Mask R-CNN in Classification and Detection of Melanoma. Comput. Methods Programs Biomed. 2021, 207, 106174. [Google Scholar] [CrossRef]
- Freudenberg, M.; Nölke, N.; Agostini, A.; Urban, K.; Wörgötter, F.; Kleinn, C. Large Scale Palm Tree Detection in High Resolution Satellite Images Using U-Net. Remote Sens. 2019, 11, 312. [Google Scholar] [CrossRef] [Green Version]
- Chu, P.; Li, Z.; Lammers, K.; Lu, R.; Liu, X. Deep learning-based apple detection using a suppression mask R-CNN. Pattern Recognit. Lett. 2021, 147, 206–211. [Google Scholar] [CrossRef]
- Loh, D.R.; Wen, X.Y.; Yapeter, J.; Subburaj, K.; Chandramohanadas, R. A Deep Learning Approach to the Screening of Malaria Infection: Automated and Rapid Cell Counting, Object Detection and Instance Segmentation Using Mask R-CNN. Comput. Med. Imaging Graph. 2021, 88, 10185. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Yuan, Z.; Peng, J.; Chen, L.; Huang, H.; Zhu, J.; Liu, Y.; Li, H. DASNet: Dual Attentive Fully Convolutional Siamese Networks for Change Detection in High-Resolution Satellite Images. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 14, 1194–1206. [Google Scholar] [CrossRef]
- Zimmermann, R.S.; Siems, J.N. Faster Training of Mask R-CNN by Focusing on Instance Boundaries. Comput. Vis. Image Underst. 2019, 188, 102795. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Qi, L.; Qin, H.; Shi, J.; Jia, J. Path Aggregation Network for Instance Segmentation. In Proceedings of the 2018, IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; Institute of Electrical and Electronics Engineers (IEEE): New York, NY, USA, 2018; pp. 8759–8768. [Google Scholar] [CrossRef] [Green Version]
- ContextCapture Software, v4.4.11; Bentley: Exton, PA, USA. Available online: https://www.bentley.com/zh/products/brands/contextcapture(accessed on 1 December 2018).
- ArcGIS Desktop Software, v10.4; ESRI: Redlands, CA, USA. Available online: https://www.esri.com/(accessed on 12 June 2021).
- VGG Image Annotator Software, v1.0; VGG: Oxford, UK. Available online: https://www.robots.ox.ac.uk/~vgg/software/via/via.html(accessed on 22 July 2017).
- Photoshop Software, Berkeley, CA, USA, v2019. Adobe. Available online: https://www.adobe.com/products/photoshop.html(accessed on 16 October 2018).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Luc, P.; Couprie, C.; Lecun, Y.; Verbeek, J. Predicting Future Instance Segmentation by Forecasting Convolutional Features. In Proceedings of the European Conference on Computer Vision, Munich, Germany, 8–14 September 2018. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Zhang, X.; Peng, C.; Cheng, D.; Sun, J. ExFuse: Enhancing Feature Fusion for Semantic Segmentation. In Proceedings of the European Conference on Computer Vision, Munich, Germany, 8–14 September 2018. [Google Scholar]
- Zhu, Q.; Du, B.; Yan, P. Boundary-Weighted Domain Adaptive Neural Network for Prostate MR Image Segmentation. IEEE Trans. Med. Imaging 2019, 99, 1. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Be, K. Topological Structural Analysis of Digitized Binary Images by Border Following. Comput. Vis. Graph. Image Process. 1985, 30, 32–46. [Google Scholar] [CrossRef]
- Shao, G.; Tang, L.; Liao, J. Overselling Overall Map Accuracy Misinforms about Research Reliability. Landsc. Ecol. 2019, 34, 2487–2492. [Google Scholar] [CrossRef] [Green Version]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015. [Google Scholar] [CrossRef] [Green Version]
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Ferreira, M.P.; Zortea, M.; Zanotta, D.C.; Shimabukuro, Y.E.; de Souza Filho, C.R. Mapping Tree Species in Tropical Seasonal Semi-Deciduous Forests with Hyperspectral and Multispectral Data. Remote Sens. Environ. 2016, 179, 66–78. [Google Scholar] [CrossRef]
- Ferreira, M.P.; Wagner, F.H.; Aragão, L.; Shimabukuro, Y.E.; Filho, C.R.S. Tree Species Classification in Tropical Forests Using Visible to Shortwave Infrared WorldView-3 Images and Texture Analysis. ISPRS J. Photogramm. Remote Sens. 2019, 149, 119–131. [Google Scholar] [CrossRef]

















 | Size | 322 mm × 242 mm × 84 mm |
| Maximum flight time | 31 min | |
| Hover precision | V: ±0.1 m; H: ±0.3 m | |
| Maximum flight speed | 72 km/h | |
| Maximum cruising mileage | 18 km | |
| Maximum wind resistance level | 5 |
| Type | Species | Field Investigation | Visual Interpretation | Similarity of Totals (%) |
|---|---|---|---|---|
| Coniferous forest | Pinus armandii | 11,776 | 11,669 | 99.09 |
| Ginkgo biloba | 15,681 | 15,552 | 99.18 | |
| Pinus tabulaeformis | 3232 | 3221 | 99.63 | |
| Broadleaf forest | Sophora japonica | 2976 | 2943 | 98.89 |
| Salix matsudana | 4408 | 4356 | 98.83 | |
| Ailanthus altissima | 10,152 | 10,045 | 98.95 | |
| Amygdalus davidiana | 2464 | 2439 | 98.98 | |
| Populus nigra | 2048 | 2030 | 99.12 | |
| Total | - | 52,737 | 52,079 | - |
| Species | Dataset | ||
|---|---|---|---|
| Train Set | Validation Set | Test Set | |
| Pinus armandii | 285 | 162 | 165 |
| Ginkgo biloba | 308 | 169 | 173 |
| Pinus tabulaeformis | 175 | 106 | 112 |
| Sophora japonica | 168 | 85 | 88 |
| Salix matsudana | 184 | 97 | 99 |
| Ailanthus altissima | 259 | 135 | 141 |
| Amygdalus davidiana | 131 | 74 | 69 |
| Populus nigra | 93 | 48 | 55 |
| Hardware | Attribute |
|---|---|
| CPU | i9-10850 |
| GPU | GTX 2080Ti 11GB |
| SSD | 1T SSD |
| Memory | 64GB |
| Type | Species | Precision (%) | Recall (%) | F1-Score (%) | Mean Average Precision (%) |
|---|---|---|---|---|---|
| Coniferous forest | Pinus armandii | 90.28 | 89.87 | 90.07 | 90.39 |
| Ginkgo biloba | 93.21 | 91.78 | 92.48 | 91.23 | |
| Pinus tabulaeformis | 92.45 | 88.71 | 90.54 | 90.14 | |
| Broadleaf forest | Sophora japonica | 80.62 | 83.42 | 81.99 | 80.72 |
| Salix matsudana | 85.44 | 82.63 | 84.01 | 83.68 | |
| Ailanthus altissima | 81.97 | 80.02 | 80.90 | 80.06 | |
| Amygdalus davidiana | 82.59 | 80.52 | 81.54 | 81.77 | |
| Populus nigra | 75.76 | 77.23 | 76.98 | 75.55 |
| Prediction Data | Reference Data | |||||||||
| Pinus armandii | Ginkgo biloba | Pinus tabulaeformis | Sophora japonica | Salix matsudana | Ailanthus altissima | Amygdalus davidiana | Populus nigra | Background | User’s accuracy | |
| Pinus armandii | 2.599 | 0.038 | 0.076 | 0.022 | 0 | 0 | 0.019 | 0 | 0.415 | 0.82 |
| Ginkgo biloba | 0.234 | 8.971 | 0.17 | 0.010 | 0 | 0.010 | 0 | 0.010 | 1.270 | 0.84 |
| Pinus tabulaeformis | 0.127 | 0.102 | 10.35 | 0.051 | 0.038 | 0.012 | 0.025 | 0.012 | 2.057 | 0.81 |
| Sophora japonica | 0.035 | 0.023 | 0.011 | 8.822 | 0.023 | 0.186 | 0.140 | 0.233 | 2.193 | 0.75 |
| Salix matsudana | 0.096 | 0.032 | 0 | 0.064 | 7.643 | 0.160 | 0.245 | 0.128 | 2.309 | 0.71 |
| Ailanthus altissima | 0.007 | 0.037 | 0.007 | 0.014 | 0.185 | 5.49 | 0.133 | 0.185 | 1.359 | 0.74 |
| Amygdalus davidiana | 0.049 | 0.037 | 0.012 | 0 | 0.111 | 0.223 | 8.949 | 0.174 | 2.858 | 0.72 |
| Populus nigra | 0.049 | 0.033 | 0.066 | 0.398 | 0.082 | 0.099 | 0.082 | 12.60 | 3.168 | 0.76 |
| Background | 0.015295 | 0.12236 | 0.1560 | 0.03 | 0.091 | 0.122 | 0.214 | 0.061 | 29.764 | 0.97 |
| Producer’s accuracy | 0.80 | 0.9 | 0.95 | 0.93 | 0.93 | 0.87 | 0.91 | 0.93 | 0.65 | - |
| Network Type | Kappa Coefficient | Overall Accuracy (%) | ||
|---|---|---|---|---|
| Training Set | Test Set | Training Set | Test Set | |
| U-net [57] | 0.75 | 0.70 | 85.42 | 81.14 |
| YOLOv3 [58] | 0.70 | 0.62 | 81.56 | 78.57 |
| Mask R-CNN [33] | 0.79 | 0.76 | 90.86 | 89.72 |
| Improved Mask R-CNN | 0.81 | 0.79 | 92.71 | 90.13 |
| Network Type | Model Parameters (million) | Time in Each Epoch (s−1) |
|---|---|---|
| U-net | 31.05 | 318 |
| YOLOv3 | 56.78 | 489 |
| Mask R-CNN | 40.76 | 372 |
| Improved Mask R-CNN | 33.47 | 327 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Zhou, J.; Wang, H.; Tan, T.; Cui, M.; Huang, Z.; Wang, P.; Zhang, L. Multi-Species Individual Tree Segmentation and Identification Based on Improved Mask R-CNN and UAV Imagery in Mixed Forests. Remote Sens. 2022, 14, 874. https://doi.org/10.3390/rs14040874
Zhang C, Zhou J, Wang H, Tan T, Cui M, Huang Z, Wang P, Zhang L. Multi-Species Individual Tree Segmentation and Identification Based on Improved Mask R-CNN and UAV Imagery in Mixed Forests. Remote Sensing. 2022; 14(4):874. https://doi.org/10.3390/rs14040874
Chicago/Turabian StyleZhang, Chong, Jiawei Zhou, Huiwen Wang, Tianyi Tan, Mengchen Cui, Zilu Huang, Pei Wang, and Li Zhang. 2022. "Multi-Species Individual Tree Segmentation and Identification Based on Improved Mask R-CNN and UAV Imagery in Mixed Forests" Remote Sensing 14, no. 4: 874. https://doi.org/10.3390/rs14040874