High-Throughput Phenotyping Analysis of Potted Soybean Plants Using Colorized Depth Images Based on A Proximal Platform
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Treatments and Measurement of Phenotypic Traits
2.2. Proximal Platform-Based Data Acquisition System
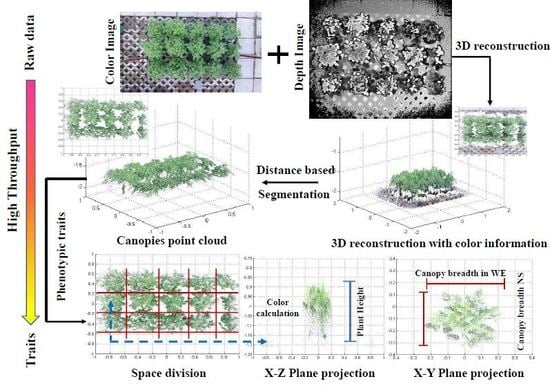
2.3. Overall Process Flow for Calculating Phenotypic Traits
2.4. Registration of Color Images
2.5. Calculation of Color Indices
2.6. Calculation of Plant Height
2.7. Calculation of Canopy Breadth
3. Results
3.1. Accuracy of Color Registration
3.2. Extraction of the Soybean Canopy Based on 3D Point Cloud
3.3. Accuracy of Plant Height
3.4. Accuracy of Canopy Breadth
3.5. Calculation of Color Indices
3.6. Comparison of Phenotypic Traits for Different Varieties
4. Discussion
4.1. Analysis of the Experimental Results
4.2. Limitation and Scalability of the Acquisition System
4.3. Future Work
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [Green Version]
- Eriksson, D.; Brinch-Pedersen, H.; Chawade, A.; Holme, I.; Hvoslef-Eide, T.A.K.; Ritala, A. Scandinavian perspectives on plant gene technology: Applications, policies and progress. Physiol. Plant. 2018, 162, 219–238. [Google Scholar] [CrossRef]
- Halewood, M.; Chiurugwi, T.; Sackville Hamilton, R.; Kurtz, B.; Marden, E.; Welch, E.; Michiels, F.; Mozafari, J.; Sabran, M.; Patron, N.; et al. Plant genetic resources for food and agriculture: Opportunities and challenges emerging from the science and information technology revolution. New Phytol. 2018, 217, 1407–1419. [Google Scholar] [CrossRef]
- Chelle, M. Phylloclimate or the Climate Perceived by Individual Plant Organs: What Is It? How to Model It? What For? New Phytol. 2005, 166, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Hawkesford, M.J.; Lorence, A. Plant phenotyping: Increasing throughput and precision at multiple scales. Funct. Plant Biol. 2017, 44, v–vii. [Google Scholar] [CrossRef]
- Celesti, M.; Tol, C.V.D.; Cogliati, S.; Panigada, C.; Yang, P.; Pinto, F.; Rascher, U.; Miglietta, F.; Colombo, R.; Rossini, M. Exploring the physiological information of sun-induced chlorophyll fluorescence through radiative transfer model inversion. Remote Sens. Environ. 2018, 215, 97–108. [Google Scholar] [CrossRef]
- Garrido, M.; Paraforos, D.S.; Reiser, D.; Vázquez Arellano, M.; Griepentrog, H.; Valero, C. 3D maize plant reconstruction based on georeferenced overlapping lidar point clouds. Remote Sens. 2015, 7, 17077–17096. [Google Scholar] [CrossRef]
- Rosell, J.R.; Llorens, J.; Sanz, R.; Jaume, A.; Ribes-Dasi, M.; Masip, J. Obtaining the three-dimensional structure of tree orchards from remote 2D terrestrial LIDAR scanning. Agric. For. Meteorol. 2009, 149, 1505–1515. [Google Scholar] [CrossRef] [Green Version]
- Goggin, F.L.; Lorence, A.; Topp, C.N. Applying high-throughput phenotyping to plant–insect interactions: Picturing more resistant crops. Curr. Opin. Insect Sci. 2015, 9, 69–76. [Google Scholar] [CrossRef]
- Großkinsky, D.K.; Pieruschka, R.; Svensgaard, J.; Uwe, R.; Svend, C.; Ulrich, S.; Thomas, R. Phenotyping in the fields: Dissecting the genetics of quantitative traits and digital farming. New Phytol. 2015, 207, 950–952. [Google Scholar] [CrossRef] [PubMed]
- Lipka, A.E.; Kandianis, C.B.; Hudson, M.E.; Yu, J.; Drnevich, J.; Bradbury, P.J. From association to prediction: Statistical methods for the dissection and selection of complex traits in plants. Curr. Opin. Plant Biol. 2015, 24, 110–118. [Google Scholar] [CrossRef]
- Stamatiadis, S.; Tsadilas, C.; Schepers, J.S. Ground-based canopy sensing for detecting effects of water stress in cotton. Plant Soil 2010, 331, 277–287. [Google Scholar] [CrossRef]
- Sun, S.; Li, C.; Paterson, A.H. In-field high-throughput phenotyping of cotton plant height using LiDAR. Remote Sens. 2017, 9, 377. [Google Scholar] [CrossRef]
- Tilly, N.; Hoffmeister, D.; Cao, Q.; Huang, Q.; Lenz-Wiedemann, S.; Miao, V.; Bareth, Y. Multitemporal crop surface models: Accurate plant height measurement and biomass estimation with terrestrial laser scanning in paddy rice. J. Appl. Remote Sens. 2014, 8, 083671. [Google Scholar] [CrossRef]
- Sritarapipat, T.; Rakwatin, P.; Kasetkasem, T. Automatic rice crop height measurement using a field server and digital image processing. Sensors 2014, 14, 900–926. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, D.; Shi, P.; Omasa, K. Estimating rice chlorophyll content and leaf nitrogen concentration with a digital still color camera under natural light. Plant Methods 2014, 10, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, H.; Li, J.; Ma, X. Recognition of soybean nutrient deficiency based on color characteristics of canopy. J. Northwest A F Univ. 2016, 44, 136–142. [Google Scholar]
- Do Amaral, E.S.; Silva, D.V.; Dos Anjos, L.; Schilling, A.C.; Dalmolin, Â.C.; Mielke, M.S. Relationships between reflectance and absorbance chlorophyll indices with RGB (Red, Green, Blue) image components in seedlings of tropical tree species at nursery stage. New For. 2018. [Google Scholar] [CrossRef]
- Mohan, P.J.; Gupta, S.D. Intelligent image analysis for retrieval of leaf chlorophyll content of rice from digital images of smartphone under natural light. Photosynthetica 2019, 57, 388–398. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, X.; Wang, H. Preliminary research on total nitrogen content prediction of sandalwood using the error-in-variable models based on digital image processing. PLoS ONE 2018, 13, e0202649. [Google Scholar] [CrossRef]
- Shi, C.; Qian, J.; Han, S.; Fan, B.; Yang, X.; Wu, X. Developing a machine vision system for simultaneous prediction of freshness indicators based on tilapia (Oreochromis niloticus) pupil and gill color during storage at 4 °C. Food Chem. 2018, 243, 134–140. [Google Scholar] [CrossRef]
- De Ocampoa, A.L.P.; Albob, J.B.; de Ocampoc, K.J. Image analysis of foliar greenness for quantifying relative plant health. Ed. Board 2015, 1, 27–31. [Google Scholar]
- Baresel, J.P.; Rischbeck, P.; Hu, Y.; Kipp, S.; Barmeier, G.; Mistele, B.; Schmidhalter, U. Use of a digital camera as alternative method for non-destructive detection of the leaf chlorophyll content and the nitrogen nutrition status in wheat. Comput. Electron. Agric. 2017, 140, 25–33. [Google Scholar] [CrossRef]
- Yadav, S.P.; Ibaraki, Y.; Gupta, S.D. Estimation of the chlorophyll content of micropropagated potato plants using RGB based image analysis. Plant Cell Tissue Organ Cult. 2010, 100, 183–188. [Google Scholar] [CrossRef]
- Padmaja, V.; Dey, M.A.K. Evaluation of leaf chlorophyll content by a non-invasive approach. Evaluation 2015, 3, 7–10. [Google Scholar]
- Sass, L.; Majer, P.; Hideg, E. Leaf hue measurements: A high-throughput screening of chlorophyll content. Methods Mol. Biol. 2012, 918, 61–69. [Google Scholar] [PubMed]
- Mishra, K.B.; Mishra, A.; Klem, K. Plant phenotyping: A perspective. Indian J. Plant Physiol. 2016, 21, 514–527. [Google Scholar] [CrossRef]
- Fiorani, F.; Schurr, U. Future scenarios for plant phenotyping. Ann. Rev. Plant Biol. 2013, 64, 267–291. [Google Scholar] [CrossRef]
- Mahlein, A.K.; Oerke, E.C.; Steiner, U.; Dehne, H.W. Recent advances in sensing plant diseases for precision crop protection. Eur. J. Plant Pathol. 2012, 133, 197–209. [Google Scholar] [CrossRef]
- Marchese, M.; Falconieri, A.; Pergola, N.; Tramutoli, V. Monitoring the Agung (Indonesia) Ash Plume of November 2017 by Means of Infrared Himawari 8 Data. Remote Sens. 2018, 6, 919. [Google Scholar] [CrossRef]
- Carlone, L.; Dong, J.; Fenu, S.; Rains, G.; Dellaert, F. Towards 4D crop analysis in precision agriculture: Estimating plant height and crown radius over time via expectation-maximization. In Proceedings of the ICRA Workshop on Robotics in Agriculture, Seattle, WA, USA, 30 May 2015. [Google Scholar]
- Kaess, M.; Johannsson, H.; Roberts, R.; Ila, V.; Leonard, J.J.; Dellaert, F. iSAM2: Incremental smoothing and mapping using the bayes tree. Int. J. Robot. Res. 2011, 31, 216–235. [Google Scholar] [CrossRef]
- Guan, H.; Liu, M.; Ma, X.; Yu, S. Three-dimensional reconstruction of soybean canopies using multisource imaging for phenotyping analysis. Remote Sens. 2018, 10, 1206. [Google Scholar] [CrossRef]
- Ma, X.; Feng, J.; Guan, H.; Liu, G. Prediction of Chlorophyll Content in Different Light Areas of Apple Tree Canopies based on the Color Characteristics of 3D Reconstruction. Remote Sens. 2018, 10, 429. [Google Scholar] [CrossRef]
- Cui, Y.; Schuon, S.; Chan, D.; Thrun, S.; Theobalt, C. 3D shape scanning with a time-of-flight camera. In Proceedings of the 2010 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), San Francisco, CA, USA, 13–18 June 2010; pp. 1173–1180. [Google Scholar]
- Andújar, D.; Dorado, J.; Fernández-Quintanilla, C.; Ribeiro, A. An Approach to the use of depth cameras for weed volume estimation. Sensors 2016, 16, 972. [Google Scholar] [CrossRef]
- Mccormick, R.F.; Truong, S.K.; Mullet, J.E. 3D sorghum reconstructions from depth images identify QTL regulating shoot architecture. Plant Physiol. 2016, 172, 823–834. [Google Scholar] [CrossRef]
- Shi, Y.; He, P.; Hu, S.; Zhang, Z.; Geng, N.; He, D. Reconstruction Method of Tree Geometric structures from point clouds based on angle-constrained space colonization algorithm. Trans. Chin. Soc. Agric. Mach. 2018, 49, 207–216. [Google Scholar]
- Yang, L.; Zhang, L.; Dong, H.; Alelaiwi, A.; Saddik, A.E. Evaluating and Improving the Depth Accuracy of Kinect for Windows v2. IEEE Sens. J. 2015, 15, 4275–4285. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, C.; Robertson, J.S.; Xu, R.; Paterson, A.H. Gphenovision: A ground mobile system with multi-modal imaging for field-based high throughput phenotyping of cotton. Sci. Rep. 2018, 8, 1213. [Google Scholar] [CrossRef]
- Bai, X.; Liu, L.; Xu, G. Colour chart histogram based supervise colour constancy. J. Tsinghua Univ. Sci. Tech. 1997, 37, 1–6. (In Chinese) [Google Scholar]
- Guo, Y.; Ge, Q.; Guo, N. Colour correction based on white balance. J. Comput. Eng. Appl. 2005, 20, 56–59. [Google Scholar]
- Muharam, F.M.; Bronson, K.F.; Maas, S.J.; Ritchie, G.L. Inter-relationships of cotton plant height, canopy width, ground cover and plant nitrogen status indicators. Field Crop. Res. 2014, 169, 58–69. [Google Scholar] [CrossRef] [Green Version]
- Peter, P.J.; Roosjen, B.B.; Juha, M.S.; Harm, M.; Bartholomeus, L.K.; Clevers, J.G.P.W. Improved estimation of leaf area index and leaf chlorophyll content of a potato crop using multi-angle spectral data—potential of unmanned aerial vehicle imagery. Int. J. Appl. Earth Obs. Geoinform. 2018, 66, 14–26. [Google Scholar]
- Taise, R.; Kunrath, G.L.; Victor, O.S.; François, G. Water use efficiency in perennial forage species: Interactions between nitrogen nutrition and water deficit. Field Crops Res. 2018, 222, 1–11. [Google Scholar]
- Rueda-Ayala, V.P.; Peña, J.M.; Höglind, M.; Bengochea-Guevara, J.M.; Andújar, D. Comparing UAV-Based Technologies and RGB-D Reconstruction Methods for Plant Height and Biomass Monitoring on Grass Ley. Sensors 2019, 19, 535. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, L.; Xiang, L.; Wu, Q.; Jiang, H. Automatic non-destructive growth measurement of leafy vegetables based on kinect. Sensors 2018, 18, 806. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Grift, T.E. A LiDAR-based crop height measurement system for Miscanthus Giganteus. Comput. Electron. Agric. 2012, 85, 70–76. [Google Scholar] [CrossRef]
- Rosell Polo, J.R.; Sanz, R.; Llorens, J.; Arnó, J.; Escolà, A.; Ribes-Dasi, M.; Masip, J.; Camp, F.; Gràcia, F.; Solanelles, F.; et al. A tractor-mounted scanning LiDAR for the non-destructive measurement of vegetative volume and surface area of tree-row plantations: A comparison with conventional destructive measurements. Biosyst. Eng. 2009, 102, 128–134. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, C.; Paterson, A.H. High throughput phenotyping of cotton plant height using depth images under field conditions. Comput. Electron. Agric. 2016, 130, 57–68. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, C.; Paterson, A.H.; Sun, S.; Xu, R.; Robert, J. Quantitative Analysis of Cotton Canopy Size in Field Conditions Using a consumer-Grade RGB-D Camera. Front. Plant Sci. 2018, 8, 2233. [Google Scholar] [CrossRef] [PubMed]
- Lachat, E.; Macher, H.; Landes, T.; Grussenmeyer, P. Assessment and calibration of a rgb-d camera (kinect v2 sensor) towards a potential use for close-range 3d modeling. Remote Sens. 2015, 7, 13070–13097. [Google Scholar] [CrossRef]
- Chhokra, P.; Chowdhury, A.; Goswami, G.; Vatsa, M.; Singh, R. Unconstrained Kinect video face database. Inform. Fusion 2018, 44, 113–125. [Google Scholar] [CrossRef]
- Mateo, F.; Soria-Olivas, E.; Carrasco, J.; Bonanad, S.; Querol, F.; Pérez-Alenda, S. HemoKinect: A Microsoft Kinect V2 Based Exergaming Software to Supervise Physical Exercise of Patients with Hemophilia. Sensors 2018, 18, 2439. [Google Scholar] [CrossRef] [PubMed]
- Timmi, A.; Coates, G.; Fortin, K.; Ackland, D.; Bryant, A.L.; Gordon, I.; Pivonka, P. Accuracy of a novel marker tracking approach based on the low-cost Microsoft Kinect v2 sensor. Med. Eng. Phys. 2018, 59, 63–69. [Google Scholar] [CrossRef]
- Zhang, Z.; Cheng, X.; Jiang, Z. Excessive saturation effect of visible light CCD. High Power Laser Part. Beams 2008, 6, 917–920. (In Chinese) [Google Scholar]
- Li, Q.; Li, G. Investigation into the Elimination of Excessive Saturation in CCD Images. Microcompu. Inform. 2010, 26, 97–99. [Google Scholar]
- Zhang, Q.; Huang, S.H.; Zhao, X.; Fu-Qi, S.I.; Zhou, H.J.; Wang, Y. The design and implementation of ccd refrigeration system of imaging spectrometer. Acta Photonica Sinica 2017, 46, 171–177. (In Chinese) [Google Scholar]
- Azadbakht, M.; Ashourloo, D.; Aghighi, H.; Radiom, S.; Alimohammadi, A. Wheat leaf rust detection at canopy scale under different LAI levels using machine learning techniques. Comput. Electron. Agric. 2019, 156, 119–128. [Google Scholar] [CrossRef]
- Vidal, T.; Gigot, C.; de Vallavieille-Pope, C.; Huber, L.; Saint-Jean, S. Contrasting plant height can improve the control of rain-borne diseases in wheat cultivar mixture: Modelling splash dispersal in 3-D canopies. Ann. Bot. 2018, 121, 1299–1308. [Google Scholar] [CrossRef]
- Singh, A.; Sah, L.P.; GC, Y.D.; Devkota, M.; Colavito, L.A.; Rajbhandari, B.P.; Muniappan, R. Evaluation of pest exclusion net to major insect pest of tomato in Kavre and Lalitpur. Nepal. J. Agric. Sci. 2018, 16, 128–137. [Google Scholar]















| 28 June | 27 July | 26 August | 5 September | |
|---|---|---|---|---|
| Light intensity/µmoles/m2/s | 753 | 529 | 845 | 477 |
| Mounting Height/m | 1.45 | 1.70 | 1.80 | 1.90 |
| Date | Under Natural Light Conditions | After Registration | Reference | Error of Registration | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R | G | B | R | G | B | R = G = B | ER | EG | EB | |
| 28 June | 143.69 | 159.85 | 175.23 | 128.75 | 129.39 | 128.94 | 128 | 0.58% | 1.09% | 0.73% |
| 27 July | 164.63 | 176.98 | 211.00 | 126.96 | 127.72 | 128.21 | 128 | 0.84% | 0.2208% | 0.16% |
| 26 August | 136.11 | 154.22 | 179.26 | 126.39 | 126.84 | 128.57 | 128 | 1.26% | 0.91% | 0.44% |
| 5 September | 176.01 | 198.54 | 225.90 | 127.22 | 127.97 | 128.96 | 128 | 0.61% | 0.03% | 0.75% |
| PH | CB in W–E | CB in N–S | h | s | i | |
|---|---|---|---|---|---|---|
| RSME | 1.92 cm | 1.23 cm | 1.85 cm | 0.0004 | 0.0018 | 0.0022 |
| MRE | 5.1% | 2.4% | 2.9% | 7.8% | 8.5% | 7.1% |
| MAE | 1.58 cm | 0.90 cm | 1.44 cm | 0.0003 | 0.0013 | 0.0013 |
| Varieties | PH | CB in W-E | CB in N-S | h | s | i |
|---|---|---|---|---|---|---|
| Kangxian 9 | 48.86 ± 2.0330 a | 53.74 ± 0.6986 a | 48.44 ± 0.5550 a | 0.0037 ± 0.0001 a | 0.0069 ± 0.0005 b | 0.9894 ± 0.0088 a |
| Fangxian 13 | 49.28 ± 1.4412 a | 50.60 ± 2.8373 a | 48.30 ± 0.4472 a | 0.0043 ± 0.0020 a | 0.0082 ± 0.0009 a | 0.9875 ± 0.0090 a |
| Fudou 6 | 49.98 ± 1.6664 a | 53.40 ± 1.3285 a | 47.80 ± 0.8367 a | 0.0035 ± 0.0007 a | 0.0059 ± 0.0006 c | 0.9906 ± 0.0016 a |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.; Zhu, K.; Guan, H.; Feng, J.; Yu, S.; Liu, G. High-Throughput Phenotyping Analysis of Potted Soybean Plants Using Colorized Depth Images Based on A Proximal Platform. Remote Sens. 2019, 11, 1085. https://doi.org/10.3390/rs11091085
Ma X, Zhu K, Guan H, Feng J, Yu S, Liu G. High-Throughput Phenotyping Analysis of Potted Soybean Plants Using Colorized Depth Images Based on A Proximal Platform. Remote Sensing. 2019; 11(9):1085. https://doi.org/10.3390/rs11091085
Chicago/Turabian StyleMa, Xiaodan, Kexin Zhu, Haiou Guan, Jiarui Feng, Song Yu, and Gang Liu. 2019. "High-Throughput Phenotyping Analysis of Potted Soybean Plants Using Colorized Depth Images Based on A Proximal Platform" Remote Sensing 11, no. 9: 1085. https://doi.org/10.3390/rs11091085