Mapping Wetland Types in Semiarid Floodplains: A Statistical Learning Approach
Abstract
:1. Introduction
- To investigate the discrimination limits of the predictive models through evaluating the performance of predictive models at three vegetation formation levels. Our approach involved building predictive models for three vegetation classification levels: at a broad functional group level (two classes: wetland vs. non-wetlands), an intermediate level (four classes) which further divides wetlands into vegetation structural groups, and a more detailed level involving vegetation floristics (dominant species—nine classes).
- To evaluate the predictive power of various predictor variables. In wetland ecosystems, vegetation distribution is strongly affected by the moisture gradient, which is often highly correlated with micro-topography [36,37]. The combined use of LiDAR-based geomorphological data and Landsat data may present an opportunity for refined vegetation mapping. Our study, therefore, assesses the individual and combined contributions of geomorphological variables to classification accuracy by building models with three combinations of predictor variables: full models with all Landsat and LiDAR DEM-based geomorphological variables; the geomorphological models with only LiDAR DEM-based geomorphological variables; and the Landsat models fitted with only Landsat variables.
2. Materials and Methods
2.1. Study Site
2.2. Wetland Vegetation Map
- (1)
- Level one contains the two broadest functional groups: flood-dependent (wetland) and non-flood-dependent terrestrial vegetation communities. PCTs were assessed as either flood-dependent (wetland) or terrestrial based on knowledge of the wetland plant indicator status of dominant plant species. The wetland indicator plant species list is from derived, descriptive information recorded in the NSW Flora Online [43].
- (2)
- Level two has three classes which divide the flood-dependent group (wetlands) into three broad structural groups: woodland, shrubland, and grassy/herbaceous wetlands. We grouped the PCTs into these structural groups based on the dominant life form of the tallest plant layer [44].
- (3)
- Level three comprises nine vegetation classes, formed by grouping PCTs per dominant species (floristics), and the conceptual understanding of the landscape position and water requirements of these communities described in the NSW Vegetation Information System Classification database [42].
2.3. Inputs of GBM Classifier
2.3.1. Geomorphological Variables
2.3.2. Variables derived from Landsat TM and ETM+ Images
- The polar angle median (V1)
- The vector distance median (V2)
- The vector distance maximum (V3)
- The hop length median (V4)
- The skewness in the vector distance based on the statistical distribution of the distance values at a pixel (V5)
2.4. Stochastic Gradient Boosting Machines
2.5. Accuracy Assessment and Model Comparison
3. Results
3.1. Model Performance
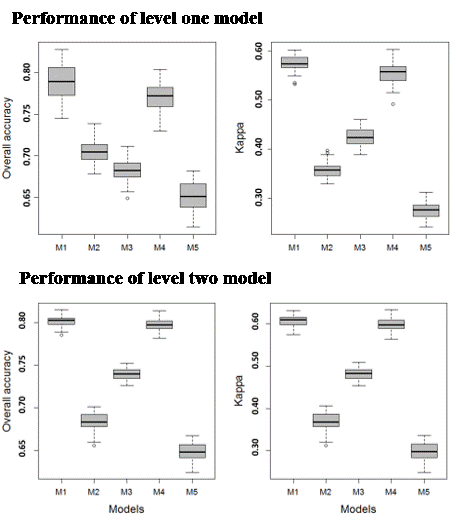
3.1.1. Level 1 Models
3.1.2. Level 2 Models
3.1.3. Level 3 Models
3.2. Relative Variable Influence
4. Discussion
4.1. The Discriminative Limits of Predictive Models Using Landsat and DEM-Derived Predictors
4.2. The Importance of Integrating Geomorphological Variables in Floodplain Vegetation Mapping
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gibbs, J.P. Wetland loss and biodiversity conservation. Conserv. Biol. 2000, 14, 314–317. [Google Scholar] [CrossRef]
- Georgiou, S.; Turner, R.K. Valuing Ecosystem Services: The Case of Multi-Functional Wetlands; Routledge: New York, NY, USA, 2012. [Google Scholar]
- Mitch, W.J.; Gosselink, J.G. Wetlands, 4th ed.; Wiley & Sons: Hoboken, NJ, USA, October 2008. [Google Scholar]
- Rebela, L.M.; Finlayson, C.M.; Nagabhatla, N. Remote Sensing and GIS for wetland inventory, mapping and change analysis. J. Environ. Manag. 2009, 90, 2144–2153. [Google Scholar] [CrossRef] [PubMed]
- Adam, E.; Mutanga, O.; Rugege, D. Multispectral and hyperspectral remote sensing for identification and mapping of wetland vegetation: A review. Wetl. Ecol. Manag. 2010, 18, 281–296. [Google Scholar] [CrossRef]
- Lehner, B.; Doll, P. Development and validation of a global database of lakes, reservoirs and wetlands. J. Hydrol. 2004, 296, le22. [Google Scholar] [CrossRef]
- Jones, K.; Lanthier, Y.; van der Voet, P.; van Valkengoed, E.; Taylor, D.; Fernandez-Prieto, D. Monitoring and assessment of wetlands using Earth Observation: The GlobWetland project. J. Environ. Manag. 2009, 90, 2154–2169. [Google Scholar] [CrossRef] [PubMed]
- Johnston, R.M.; Barson, M.M. Remote sensing of Australian wetlands: An evaluation of Landsat TM data for inventory and classification. Mar. Freshw. Res. 1993, 44, 235–252. [Google Scholar] [CrossRef]
- Merot, P.; Squividant, H.; Aurousseau, P.; Hefting, M.; Burt, T.; Maitre, V.; Kruk, M.; Butturini, A.; Thenail, C.; Viaud, V. Testing a climato-topographic indiex for predicting wetlands distribution along a European climate gradient. Ecol. Model. 2003, 163, 51–71. [Google Scholar] [CrossRef]
- Curie, F.; Gaillard, S.; Ducharne, A.; Bendjoudi, H. Geomorphological methods to characterise wetlands at the scale of the Seine watershed. Sci. Total Environ. 2007, 375, 59–68. [Google Scholar] [CrossRef]
- MacAlister, C.; Mahaxay, M. Mapping wetlands in the Lower Mekong Basin for wetland resource and conservation management using Landsat ETM images and field survey data. J. Environ. Manag. 2009, 90, 2130–2137. [Google Scholar] [CrossRef]
- Finlayson, C.M.; Davidson, N.C.; Spiers, A.G.; Stevenson, N.J. Global wetland inventory–current status and future priorities. Mar. Freshw. Res. 1999, 50, 717–727. [Google Scholar] [CrossRef]
- Zedler, J.B.; Kercher, S. Wetland resources: Status, trends, ecosystem services, and restorability. Annu. Rev. Environ. Resour. 2005, 30, 39–74. [Google Scholar] [CrossRef]
- Brinson, M.M.; Bradshaw, H.D.; Kane, E.S. Nutrient assimilative capacity of an alluvial floodplain swamp. J. Appl. Ecol. 1984, 21, 1041–1057. [Google Scholar] [CrossRef]
- Bernal, B.; Mitsch, W.J. Comparing carbon sequestration in temperate freshwater wetland communities. Glob. Chang. Biol. 2012, 18, 1636–1647. [Google Scholar] [CrossRef]
- Maltby, E.; Barker, T. (Eds.) The Wetlands Handbook, 2 Volume Set; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- Zedler, J.B. Progress in wetland restoration ecology. Trends Ecol. Evol. 2000, 15, 402–407. [Google Scholar] [CrossRef]
- Capon, S.J.; Brock, M.A. Flooding, soil seed bank dynamics and vegetation resilience of a hydrologically variable desert floodplain. Freshw. Biol. 2006, 51, 206–223. [Google Scholar] [CrossRef] [Green Version]
- Follner, K.; Henle, K. The performance of plants, molluscs, and carabid beetles as indicators of hydrological conditions in floodplain grasslands. Int. Rev. Hydrobiol. 2006, 91, 364–379. [Google Scholar] [CrossRef]
- Honsova, D.; Hejcman, M.; Klaudisova, M.; Pavlu, V.; Kocourkova, D.; Hakl, J. Species composition of an alluvial meadow after 40 years of applying nitrogen, phospohorus and potassium fertilizer. Preslia 2007, 79, 245–258. [Google Scholar]
- Junk, W.J.; Piedade, M.T.F.; Schöngart, J.; Cohn-Haft, M.; Adeney, J.M.; Wittmann, F.A. Classification of major naturally-occurring Amazonian lowland wetlands. Wetlands 2011, 31, 623–640. [Google Scholar] [CrossRef]
- Küchler, A.W. The classification of vegetation. In Vegetation Mapping; Springer: Dordrecht, The Netherlands, 1988; pp. 67–80. [Google Scholar]
- Ridgeway, G. Package ‘GBM’: Generalized Boosted Regression Models; R Package Version 2.1.3; Scientific Research: Wuhan, China, 2013. [Google Scholar]
- Schmidt, K.S.; Skidmore, A.K. Spectral discrimination of vegetation types in a coastal wetland. Remote Sens. Environ. 2003, 85, 92–108. [Google Scholar] [CrossRef]
- Shaw, G.A.; Burke, H.K. Spectral imaging for remote sensing. Linc. Lab. J. 2003, 14, 3–28. [Google Scholar]
- Zhang, C.; Selch, D.; Cooper, H. A Framework to Combine Three Remotely Sensed Data Sources for Vegetation Mapping in the Central Florida Everglades. Wetlands 2016, 36, 201–213. [Google Scholar] [CrossRef]
- Gómez-Chova, L.; Tuia, D.; Moser, G.; Camps-Valls, G. Multimodal classification of remote sensing images: A review and future directions. IEEE 2015, 103, 1560–1584. [Google Scholar] [CrossRef]
- Joshi, N.; Baumann, M.; Ehammer, A.; Fensholt, R.; Grogan, K.; Hostert, P.; Jepsen, M.R.; Kuemmerle, T.; Meyfroidt, P.; Mitchard, E.T.; et al. A Review of the Application of Optical and Radar Remote Sensing Data Fusion to Land Use Mapping and Monitoring. Remote Sens. 2016, 8, 70. [Google Scholar] [CrossRef]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning; Springer: New York, NY, USA, 2013; Volume 6. [Google Scholar]
- van Beijma, S.; Comber, A.; Lamb, A. Random forest classification of salt marsh vegetation habitats using quad-polarimetric airborne SAR, elevation and optical RS data. Remote Sens. Environ. 2014, 149, 118–129. [Google Scholar] [CrossRef]
- Szantoi, Z.; Escobedo, F.J.; Abd-Elrahman, A.; Pearlstine, L.; Dewitt, B.; Smith, S. Classifying spatially heterogeneous wetland communities using machine learning algorithms and spectral and textural features. Environ. Monit. Assess. 2015, 187, 1–15. [Google Scholar] [CrossRef]
- Lang, M.; McCarty, G.; Oesterling, R.; Yeo, I.Y. Topographic metrics for improved mapping of forested wetlands. Wetlands 2013, 33, 141–155. [Google Scholar] [CrossRef]
- Maxa, M.; Bolstad, P. Mapping northern wetlands with high resolution satellite images and LiDAR. Wetlands 2009, 29, 248. [Google Scholar] [CrossRef]
- Rampi, L.P.; Knight, J.F.; Pelletier, K.C. Wetland mapping in the upper midwest United States. Photogramm. Eng. Remote Sens. 2014, 80, 439–448. [Google Scholar] [CrossRef]
- Tang, Z.; Li, R.; Li, X.; Jiang, W.; Hirsh, A. Capturing LiDAR-Derived Hydrologic Spatial Parameters to Evaluate Playa Wetlands. J. Am. Water Resour. Assoc. 2014, 50, 234–245. [Google Scholar] [CrossRef]
- Fisk, M.C.; Schmidt, S.K.; Seastedt, T.R. Topographic patterns of above-and belowground production and nitrogen cycling in alpine tundra. Ecology 1998, 79, 2253–2266. [Google Scholar] [CrossRef]
- Zelnik, I.; Čarni, A. Distribution of plant communities, ecological strategy types and diversity along a moisture gradient. Community Ecology 2008, 9, 1–9. [Google Scholar] [CrossRef]
- Australian Bureau of Meteorology (BOM). Climate Statistics for Australian Locations. 2008. Available online: http://www.bom.gov.au/climate/averages/tables/cw_070282.shtml (accessed on 26 April 2017).
- Ecological Australia. Vegetation of the Barwon-Darling and Condamine-Balonne Floodplain Systems of New South. Wales: Mapping and Survey of Plant Community Types. Prepared for the Murray Darling Basin Authority; 2015. Available online: https://www.mdba.gov.au/sites/default/files/pubs/MDBA_vegetation_mapping_report_final.pdf (accessed on 12 December 2018).
- Schultz, N.; Gowans, S.; Westbrooke, M. Survey and Mapping of Darling Floodplain Vegetation between Tilpa and Brewarrina; Prepared by the NSW Government Office of Environment and Heritage by Centre for Environmental Management; Federation University Australia: Ballarat, Australia, 2014. [Google Scholar]
- Gowans, S.; Milne, R.; Westbrooke, M.; Palmer, G. Survey of Vegetation and Vegetation Condition of Toorale; Prepared for the NSW Government Office for Environment and Heritage by the Centre for Environmental Management; University of Ballarat: Ballarat, Victoria, Australia, 2012. [Google Scholar]
- OEH, New South Wales. Vegetation Information System: Classification. 2011. Available online: http://www.environment.nsw.gov.au/research/Visclassification.htm (accessed on 17 October 2016).
- PlantNET (The NSW Plant Information Network System). Royal Botanic Gardens and Domain Trust, Sydney. Available online: http://plantnet.rbgsyd.nsw.gov.au (accessed on 5 March 2017).
- Specht, R.L. Vegetation. In Australian Environment, 4th ed.; Leeper, G.W., Ed.; Melbourne University Press: Melbourne, Australia, 1970; pp. 44–67. [Google Scholar]
- Geoscience Australia. Digital Elevation Model (DEM) of Australia Derived from LiDAR 5 Metre Grid. 2016. Available online: http://www.ga.gov.au/metadata-gateway/metadata/record/gcat_89644 (accessed on 24 June 2016).
- Singh, K.K.; Vogler, J.B.; Shoemaker, D.A.; Meentemeyer, R.K. LiDAR-Landsat data fusion for large-area assessment of urban land cover: Balancing spatial resolution, data volume and mapping accuracy. ISPRS J. Photogramm. Remote Sens. 2012, 74, 110–121. [Google Scholar] [CrossRef]
- Moore, I.D.; Gessler, P.E.; Nielsen, G.A.E.; Peterson, G.A. Soil attribute prediction using terrain analysis. Soil Sci. Soc. Am. J. 1993, 57, 443–452. [Google Scholar] [CrossRef]
- McNab, W.H. A topographic index to quantify the effect of mesoscale landform on site productivity. Can. J. For. Res. 1993, 23, 1100–1107. [Google Scholar] [CrossRef]
- Martinez, J.M.; Le Toan, T. Mapping of flood dynamics and spatial distribution of vegetation in the Amazon floodplain using multitemporal SAR data. Remote Sens. Environ. 2007, 108, 209–223. [Google Scholar] [CrossRef]
- Fisher, A.; Flood, N.; Danaher, T. Comparing Landsat water index methods for automated water classification in eastern Australia. Remote Sens. Environ. 2016, 175, 167–182. [Google Scholar] [CrossRef]
- Harvey, K.R.; Hill, G.J.E. Vegetation mapping of a tropical freshwater swamp in the Northern Territory, Australia: A comparison of aerial photography, Landsat TM and SPOT satellite imagery. Int. J. Remote Sens. 2001, 22, 2911–2925. [Google Scholar] [CrossRef] [Green Version]
- Dronova, I.; Gong, P.; Wang, L.; Zhong, L. Mapping dynamic cover types in a large seasonally flooded wetland using extended principal component analysis and object-based classification. Remote Sens. Environ. 2015, 158, 193–206. [Google Scholar] [CrossRef]
- Zhao, Y.; Feng, D.; Yu, L.; Wang, X.; Chen, Y.; Bai, Y.; Hernández, H.J.; Galleguillos, M.; Estades, C.; Biging, G.S.; et al. Detailed dynamic land cover mapping of Chile: Accuracy improvement by integrating multi-temporal data. Remote Sens. Environ. 2016, 183, 170–185. [Google Scholar] [CrossRef]
- Flood, N.; Danaher, T.; Gill, T.; Gillingham, S. An operational scheme for deriving standardised surface reflectance from Landsat TM/ETM+ and SPOT HRG imagery for Eastern Australia. Remote Sens. 2013, 5, 83–109. [Google Scholar] [CrossRef]
- Flood, N. Seasonal composite Landsat TM/ETM+ images using the Medoid (a multi-dimensional median). Remote Sens. 2013, 5, 6481–6500. [Google Scholar] [CrossRef]
- Ridgeway, G. Generalized Boosted Models: A Guide to the Gbm Package. 2007. Available online: http://cran.open-source-solution.org/web/packages/gbm/vignettes/gbm.PDF (accessed on 22 November 2018).
- Elith, J.; Leathwick, J.R.; Hastie, T. A working guide to boosted regression trees. J. Anim. Ecol. 2008, 77, 802–813. [Google Scholar] [CrossRef] [Green Version]
- Hastie, T.; Tibshirani, R.; Friedman, J.H. The Elements of Statistical Learning; Springer: New York, NY, USA, 2009. [Google Scholar]
- Breiman, L.; Freidman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; Wadsworth: Monterey, CA, USA, 1984. [Google Scholar]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Fay, M.P.; Shaw, P.A. Exact and Asymptotic Weighted Logrank Tests for Interval Censored Data: The interval R package. J. Stat. Softw. 2010, 36, 1–34. [Google Scholar] [CrossRef]
- Proschan, M.; Glimm, E.; Posch, M. Connections between permutation and t-tests: Relevance to adaptive methods. Stat. Med. 2014, 33, 4734–4742. [Google Scholar] [CrossRef]
- McNemar, Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika 1947, 12, 153–157. [Google Scholar] [CrossRef]
- Duro, D.C.; Franklin, S.E.; Dube, M.G. A comparison of pixel-based and object-based image analysis using selected machine learning algorithms for the classification of agricultural landscapes using SPOT-5 HRG imagery. Remote Sens. Environ. 2012, 118, 259–272. [Google Scholar] [CrossRef]
- Schmidt, T.; Schuster, C.; Kleinschmit, B.; Förster, M. Evaluating an Intra-Annual Time Series for Grassland Classification—How Many Acquisitions and What Seasonal Origin Are Optimal? IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 3428–3439. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef]
- Pearce, J.; Ferrier, S. Evaluating the predictive performance of habitat models developed using logistic regression. Ecol. Model. 2000, 133, 225–245. [Google Scholar] [CrossRef] [Green Version]
- Kayastha, N. Application on Lidar and Time Series Landsat Data for Mapping and Monitoring Wetlands. Ph.D. Dissertation, Virginia Polytechnic Institute, Blacksburg, VA, USA, 2014. [Google Scholar]
- Corcoran, J.M.; Knight, J.F.; Gallant, A.L. Influence of multi-source and multi-temporal remotely sensed and ancillary data on the accuracy of random forest classification of wetlands in Northern Minnesota. Remote Sens. 2013, 5, 3212–3238. [Google Scholar] [CrossRef]
- Wen, L.; Macdonald, R.; Morrison, T.; Hameed, T.; Saintilan, N.; Ling, J. From hydrodynamic to hydrological modelling: Investigating long-term hydrological regimes of key wetlands in the Macquarie Marshes, a semi-arid lowland floodplain in Australia. J. Hydrol. 2013, 500, 45–61. [Google Scholar] [CrossRef]
- Chawla, N.V.; Japkowicz, N.; Kotcz, A. Editorial: Special issue on learning from imbalanced data sets. SIGKDD Explor. 2004, 6, 1–6. [Google Scholar] [CrossRef]
- Provost, F. Machine learning from imbalanced data sets 101. In Proceedings of the AAAI’2000 Workshop on Imbalanced Data Sets, Austin, TX, USA, 30 July–1 August 2000; pp. 1–3. [Google Scholar]
- Sims, N.C.; Colloff, M.J. Remote sensing of vegetation responses to flooding of a semi-arid floodplain: Implications for monitoring ecological effects of environmental flows. Ecol. Ind. 2012, 18, 387–391. [Google Scholar] [CrossRef]
- Brock, P.M. The significance of the physical environment of the Macquarie Marshes. Aust. Geogr. 1998, 29, 71–90. [Google Scholar] [CrossRef]
- Hestir, E.L.; Brando, V.E.; Bresciani, M.; Giardino, C.; Matta, E.; Villa, P.; Dekker, A.G. Measuring freshwater aquatic ecosystems: The need for a hyperspectral global mapping satellite mission. Remote Sens. Environ. 2015, 167, 181–195. [Google Scholar] [CrossRef] [Green Version]
- Vaiphasa, C.; Ongsomwang, S.; Vaiphasa, T.; Skidmore, A. Tropical mangrove species discrimination using hyperspectral data: A laboratory study. Estuar. Coast. Shelf Sci. 2005, 65, 371–379. [Google Scholar] [CrossRef]
- Mertes, L.A.; Daniel, D.L.; Melack, J.M.; Nelson, B.; Martinelli, L.A.; Forsberg, B.R. Spatial patterns of hydrology, geomorphology, and vegetation on the floodplain of the Amazon River in Brazil from aremotesensing perspective. Geomorphology 1995, 13, 215–232. [Google Scholar] [CrossRef]
- Wright, C.; Gallant, A. Improved wetland remote sensing in Yellowstone National Park using classification trees to combine TM imagery and ancillary environmental data. Remote Sens. Environ. 2007, 107, 582–605. [Google Scholar] [CrossRef]
- Knight, J.F.; Tolcser, B.P.; Corcoran, J.M.; Rampi, L.P. The effects of data selection and thematic detail on the accuracy of high spatial resolution wetland classifications. Photogramm. Eng. Remote Sens. 2013, 79, 613–623. [Google Scholar] [CrossRef]
- Zlinszky, A.; Mücke, W.; Lehner, H.; Briese, C.; Pfeifer, N. Categorizing wetland vegetation by airborne laser scanning on Lake Balaton and Kis-Balaton, Hungary. Remote Sens. 2012, 4, 1617–1650. [Google Scholar] [CrossRef]
- Maxwell, A.E.; Warner, T.A.; Strager, M.P. Predicting palustrine wetland probability using random forest machine learning and digital elevation data-derived terrain variables. Photogramm. Eng. Remote Sens. 2016, 82, 437–447. [Google Scholar] [CrossRef]
- Wilson, J.P.; Gallant, J.C. Digital terrain analysis. Terrain Anal. Princ. Appl. 2000, 6, 1–27. [Google Scholar]
- Maxwell, A.E.; Warner, T.A. Is high spatial resolution DEM data necessary for mapping palustrine wetlands? Int. J. Remote Sens. 2019, 40, 118–137. [Google Scholar] [CrossRef]
- McGarigal, K.; Tagil, S.; Cushman, S. Surfacemetrics: An alternative to patchmetrics for the quantification of landscape structure. Landsc. Ecol. 2009, 24, 433–450. [Google Scholar] [CrossRef]
- Wen, L.; Powell, M.; Saintilan, N. Landscape position strongly affects the resistance and resilience to water deficit anomaly of floodplain vegetation community. Ecohydrology 2018, 11, e2027. [Google Scholar] [CrossRef]






| Level 1 | Level 2 | Level 3 | Area (ha) | Sampled Points |
|---|---|---|---|---|
| L1: Wetlands | L11: Woody wetlands | L111: River red gum forest | 8671 | 2560 |
| L112: Black box woodland | 14,082 | 4096 | ||
| L113: Coolibah woodland | 179,879 | 19,465 | ||
| L114: Other woodland | 16,132 | 1750 | ||
| L12: Shrub dominated wetlands | L121: Lignum shrubland | 24,852 | 4601 | |
| L122: Nitre Goosefoot shrubland | 326 | 106 | ||
| L123: Other Shrublands | 411 | 233 | ||
| L13: Grassy wetlands | L131: Inland Floodplain Swamp | 2648 | 820 | |
| L2: Terrestrial | L21: Terrestrial | L211: Terrestrial | 291,740 | 23,374 |
| Model | L1 | L2 | BA 3 | OAA 4 | Kappa | AUC 5 | Verdict 6 | ||
|---|---|---|---|---|---|---|---|---|---|
| UA 1 | PA 2 | UA | PA | ||||||
| M1 | 83.54 | 88.63 | 83.96 | 77.30 | 82.97 | 83.73 | 0.64 | 0.80 | Substantial |
| M2 | 75.37 | 80.75 | 72.42 | 65.70 | 73.22 | 74.20 | 0.44 | 0.70 | Moderate |
| M3 | 78.92 | 84.33 | 77.64 | 70.72 | 77.55 | 78.41 | 0.54 | 0.75 | Moderate |
| M4 | 82.82 | 88.81 | 83.95 | 76.07 | 82.44 | 83.27 | 0.63 | 0.80 | Substantial |
| M5 | 68.21 | 61.16 | 72.32 | 78.07 | 69.64 | 70.72 | 0.37 | 0.67 | Fair |
| Model | L11 | L12 | L13 | L21 | OAA | Kappa | AUC | Verdict | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UA | PA | BA | UA | PA | BA | UA | PA | BA | UA | PA | BA | |||||
| M1 | 82.80 | 83.89 | 83.63 | 69.39 | 72.69 | 84.03 | 77.36 | 70.66 | 85.31 | 78.95 | 76.44 | 82.06 | 79.57 | 0.75 | 76.12 | Substantial |
| M2 | 70.93 | 75.78 | 73.06 | 58.75 | 55.31 | 74.85 | 69.23 | 66.88 | 83.39 | 66.93 | 62.37 | 71.86 | 68.04 | 0.55 | 64.18 | Moderate |
| M3 | 78.32 | 77.48 | 78.50 | 52.45 | 61.70 | 76.78 | 55.58 | 34.43 | 67.17 | 69.94 | 67.17 | 74.85 | 69.30 | 0.62 | 67.97 | Moderate |
| M4 | 82.43 | 83.55 | 83.28 | 68.38 | 72.09 | 83.64 | 92.21 | 57.45 | 78.75 | 77.90 | 75.58 | 81.31 | 78.92 | 0.74 | 75.83 | Substantial |
| M5 | 67.93 | 71.83 | 69.72 | 52.99 | 53.39 | 73.26 | 70.82 | 54.81 | 77.38 | 62.15 | 57.60 | 68.19 | 64.00 | 0.49 | 61.06 | Fair |
| Classes | M1 | M2 | M3 | M4 | M5 | |
|---|---|---|---|---|---|---|
| L111 | UA | 79.23 | 72.58 | 68.25 | 78.77 | 56 |
| PA | 75.16 | 72.86 | 62.43 | 70.79 | 56.22 | |
| BA | 87.24 | 85.96 | 80.73 | 85.06 | 77.34 | |
| L112 | UA | 58.18 | 50.23 | 60.15 | 71.98 | 42.14 |
| PA | 38.84 | 33.00 | 21.44 | 34.86 | 35.92 | |
| BA | 69.14 | 66.17 | 60.62 | 67.27 | 67.48 | |
| L113 | UA | 63.73 | 52.47 | 57.45 | 62.84 | 48.7 |
| PA | 65.95 | 57.29 | 59.21 | 66.11 | 51.91 | |
| BA | 66.87 | 57.44 | 61.01 | 66.47 | 53.47 | |
| L114 | UA | 51.88 | 43.93 | 53.85 | 65.68 | 35.84 |
| PA | 32.54 | 26.7 | 15.14 | 28.56 | 29.62 | |
| BA | 62.84 | 59.87 | 54.32 | 60.97 | 61.18 | |
| L121 | UA | 67.84 | 71.08 | 54.81 | 91.5 | 67.52 |
| PA | 73.58 | 69.74 | 33.89 | 57.29 | 55.02 | |
| BA | 84.39 | 75.68 | 76.89 | 84.19 | 73.7 | |
| L122 | UA | 56.33 | 39.71 | 32.73 | 51.37 | 32.25 |
| PA | 39.38 | 25.46 | 16.66 | 35.12 | 19.21 | |
| BA | 63.86 | 56.85 | 52.45 | 61.71 | 53.71 | |
| L123 | UA | 52.15 | 35.53 | 28.55 | 47.19 | 28.07 |
| PA | 35.20 | 21.28 | 12.48 | 30.94 | 15.03 | |
| BA | 59.68 | 52.67 | 48.27 | 57.53 | 49.53 | |
| L131 | UA | 78.21 | 58.84 | 51.61 | 67.64 | 51.38 |
| PA | 72.01 | 56.94 | 61.67 | 73.24 | 54.44 | |
| BA | 85.91 | 84.75 | 66.82 | 78.59 | 77.39 | |
| L211 | UA | 78.67 | 67.06 | 69.53 | 77.72 | 61.92 |
| PA | 76.06 | 63.63 | 66.82 | 75.28 | 58.1 | |
| BA | 81.77 | 72.32 | 74.53 | 81.08 | 68.2 | |
| OAA | 76.95 | 66.15 | 67.95 | 76.36 | 60.8 | |
| Kappa | 0.57 | 0.4 | 0.43 | 0.56 | 0.32 | |
| AUC | 0.65 | 0.6 | 0.56 | 0.64 | 0.53 | |
| Verdict | Substantial | Moderate | Moderate | Substantial | Fair | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Powell, M.; Hodgins, G.; Danaher, T.; Ling, J.; Hughes, M.; Wen, L. Mapping Wetland Types in Semiarid Floodplains: A Statistical Learning Approach. Remote Sens. 2019, 11, 609. https://doi.org/10.3390/rs11060609
Powell M, Hodgins G, Danaher T, Ling J, Hughes M, Wen L. Mapping Wetland Types in Semiarid Floodplains: A Statistical Learning Approach. Remote Sensing. 2019; 11(6):609. https://doi.org/10.3390/rs11060609
Chicago/Turabian StylePowell, Megan, Grant Hodgins, Tim Danaher, Joanne Ling, Michael Hughes, and Li Wen. 2019. "Mapping Wetland Types in Semiarid Floodplains: A Statistical Learning Approach" Remote Sensing 11, no. 6: 609. https://doi.org/10.3390/rs11060609