Arctic Vegetation Mapping Using Unsupervised Training Datasets and Convolutional Neural Networks
Abstract
:1. Introduction
2. Study Area
3. Materials and Methods
3.1. Geospatial and In Situ Datasets
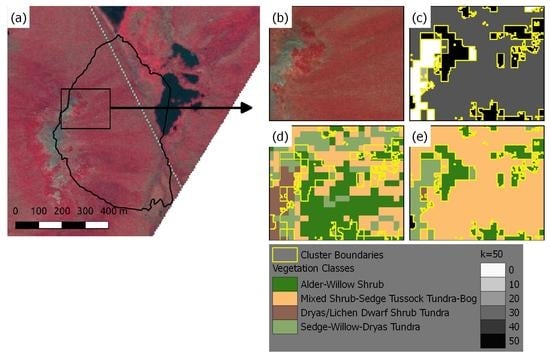
3.2. Unsupervised Classification Based Vegetation Mapping (UCVM)
3.3. Convolutional Neural Network Models for Vegetation Mapping
3.4. Validation of Vegetation Maps
4. Results
4.1. Development and Evaluation of UCVM Maps
4.2. Analysis of Datasets
4.3. CNN Based Vegetation Maps
4.3.1. Training CNN Models
4.3.2. CNN Models Trained Using UCVM
4.3.3. CNN Models Trained with AKEVT
4.3.4. Summary of Best Performing Models
5. Discussion
5.1. Vegetation Classification Trends
5.2. Remote Sensing Datasets & Multisensor Fusion
5.3. Training Label Generation
5.4. CNN Model Architecture
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Overland, J.E.; Wang, M.; Walsh, J.E.; Stroeve, J.C. Future Arctic climate changes: Adaptation and mitigation time scales. Earth’s Future 2014, 2, 68–74. [Google Scholar] [CrossRef] [Green Version]
- Lader, R.; Walsh, J.E.; Bhatt, U.S.; Bieniek, P.A. Projections of Twenty-First-Century Climate Extremes for Alaska via Dynamical Downscaling and Quantile Mapping. J. Appl. Meteorol. Climatol. 2017, 56, 2393–2409. [Google Scholar] [CrossRef]
- Zhang, W.; Miller, P.A.; Smith, B.; Wania, R.; Koenigk, T.; Döscher, R. Tundra shrubification and tree-line advance amplify arctic climate warming: results from an individual-based dynamic vegetation model. Environ. Res. Lett. 2013, 8, 034023. [Google Scholar] [CrossRef] [Green Version]
- Tranvik, L. Carbon cycling in the Arctic. Science 2014, 345, 870. [Google Scholar] [CrossRef] [PubMed]
- Pearson, R.G.; Phillips, S.J.; Loranty, M.M.; Beck, P.S.A.; Damoulas, T.; Knight, S.J.; Goetz, S.J. Shifts in Arctic vegetation and associated feedbacks under climate change. Nat. Clim. Chang. 2013, 3, 673–677. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, D.M.; Swenson, S.C. Permafrost response to increasing Arctic shrub abundance depends on the relative influence of shrubs on local soil cooling versus large-scale climate warming. Environ. Res. Lett. 2011, 6, 045504. [Google Scholar] [CrossRef] [Green Version]
- McGuire, A.D.; Koven, C.; Lawrence, D.M.; Clein, J.S.; Xia, J.; Beer, C.; Burke, E.; Chen, G.; Chen, X.; Delire, C.; et al. Variability in the sensitivity among model simulations of permafrost and carbon dynamics in the permafrost region between 1960 and 2009. Glob. Biogeochem. Cycles 2016, 30, 1015–1037. [Google Scholar] [CrossRef] [Green Version]
- Rupp, T.S.; Chapin, F.S.; Starfield, A.M. Response of subarctic vegetation to transient climatic change on the Seward Peninsula in north-west Alaska. Glob. Chang. Biol. 2000, 6, 541–555. [Google Scholar] [CrossRef] [Green Version]
- Higuera, P.; Peters, M.; Brubaker, L.; Gavin, D. Frequent Fires in Ancient Shrub Tundra: Implications of Paleorecords for Arctic Environmental Change. PLoS ONE 2008, 3, e0001744. [Google Scholar] [CrossRef]
- Tang, G.; Yuan, F.; Bisht, G.; Hammond, G.E.; Lichtner, P.C.; Kumar, J.; Mills, R.T.; Xu, X.; Andre, B.; Hoffman, F.M.; et al. Addressing Numerical Challenges in Introducing a Reactive Transport Code into a Land Surface Model: A Biogeochemical Modeling Proof-of-concept with CLM–PFLOTRAN 1.0. Geosci. Model Dev. 2016, 9, 927–946. [Google Scholar] [CrossRef]
- Bisht, G.; Huang, M.; Zhou, T.; Chen, X.; Dai, H.; Hammond, G.; Riley, W.; Downs, J.; Liu, Y.; Zachara, J. Coupling a three-dimensional subsurface flow and transport model with a land surface model to simulate stream-aquifer-land interactions (PFLOTRAN_CLM v1.0). Geosci. Model Dev. Discuss. 2017, 10, 4539–4562. [Google Scholar] [CrossRef]
- Langford, Z.; Kumar, J.; Hoffman, F.M.; Norby, R.J.; Wullschleger, S.D.; Sloan, V.L.; Iversen, C.M. Mapping Arctic Plant Functional Type Distributions in the Barrow Environmental Observatory Using WorldView-2 and LiDAR Datasets. Remote Sens. 2016, 8, 733. [Google Scholar] [CrossRef]
- Lindsay, C.; Zhu, J.; Miller, A.E.; Kirchner, P.; Wilson, T.L. Deriving Snow Cover Metrics for Alaska from MODIS. Remote Sens. 2015, 7, 12961–12985. [Google Scholar] [CrossRef] [Green Version]
- Macander, M.J.; Frost, G.V.; Nelson, P.R.; Swingley, C.S. Regional Quantitative Cover Mapping of Tundra Plant Functional Types in Arctic Alaska. Remote Sens. 2017, 9, 1024. [Google Scholar] [CrossRef]
- Verbyla, D.; Hegel, T.; Nolin, A.W.; van de Kerk, M.; Kurkowski, T.A.; Prugh, L.R. Remote Sensing of 2000–2016 Alpine Spring Snowline Elevation in Dall Sheep Mountain Ranges of Alaska and Western Canada. Remote Sens. 2017, 9, 1157. [Google Scholar] [CrossRef]
- Bratsch, S.N.; Epstein, H.E.; Buchhorn, M.; Walker, D.A. Differentiating among Four Arctic Tundra Plant Communities at Ivotuk, Alaska Using Field Spectroscopy. Remote Sens. 2016, 8, 51. [Google Scholar] [CrossRef]
- Davidson, S.J.; Santos, M.J.; Sloan, V.L.; Watts, J.D.; Phoenix, G.K.; Oechel, W.C.; Zona, D. Mapping Arctic Tundra Vegetation Communities Using Field Spectroscopy and Multispectral Satellite Data in North Alaska, USA. Remote Sens. 2016, 8, 978. [Google Scholar] [CrossRef]
- Schmitt, M.; Zhu, X.X. Data Fusion and Remote Sensing: An ever-growing relationship. IEEE Geosci. Remote Sens. Mag. 2016, 4, 6–23. [Google Scholar] [CrossRef]
- Chen, Y.; Li, C.; Ghamisi, P.; Jia, X.; Gu, Y. Deep Fusion of Remote Sensing Data for Accurate Classification. IEEE Geosci. Remote Sens. Lett. 2017, 14, 1253–1257. [Google Scholar] [CrossRef]
- Hargrove, W.W.; Hoffman, F.M.; Hessburg, P.F. Mapcurves: A Quantitative Method for Comparing Categorical Maps. J. Geogr. Syst. 2006, 8, 187–208. [Google Scholar] [CrossRef]
- Bond-Lamberty, B.; Epron, D.; Harden, J.; Harmon, M.E.; Hoffman, F.M.; Kumar, J.; McGuire, A.D.; Vargas, R. Estimating Heterotrophic Respiration at Large Scales: Challenges, Approaches, and Next Steps. Ecosphere 2016, 7, e01380. [Google Scholar] [CrossRef]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, L.; Du, B. Deep Learning for Remote Sensing Data: A Technical Tutorial on the State of the Art. IEEE Geosci. Remote Sens. Mag. 2016, 4, 22–40. [Google Scholar] [CrossRef]
- Langford, Z.L.; Kumar, J.; Hoffman, F.M. Convolutional Neural Network Approach for Mapping Arctic Vegetation using Multi-Sensor Remote Sensing Fusion. In Proceedings of the 2017 IEEE International Conference on Data Mining Workshops (ICDMW 2017), New Orleans, LA, USA, 18–21 November 2017; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA; Conference Publishing Services (CPS): Washington, DC, USA, 2017. [Google Scholar] [CrossRef]
- Xie, W.; Li, Y. Hyperspectral Imagery Denoising by Deep Learning With Trainable Nonlinearity Function. IEEE Geosci. Remote Sens. Lett. 2017, 14, 1963–1967. [Google Scholar] [CrossRef]
- Xu, X.; Li, W.; Ran, Q.; Du, Q.; Gao, L.; Zhang, B. Multisource Remote Sensing Data Classification Based on Convolutional Neural Network. IEEE Trans. Geosci. Remote Sens. 2018, 56, 937–949. [Google Scholar] [CrossRef]
- Hunt, S.; Yu, Z.; Jones, M. Lateglacial and Holocene climate, disturbance and permafrost peatland dynamics on the Seward Peninsula, western Alaska. Quat. Sci. Rev. 2013, 63, 42–58. [Google Scholar] [CrossRef]
- Silapaswan, C.; Verbyla, D.; McGuire, A. Land Cover Change on the Seward Peninsula: The Use of Remote Sensing to Evaluate the Potential Influences of Climate Warming on Historical Vegetation Dynamics. Can. J. Remote Sens. 2001, 27, 542–554. [Google Scholar] [CrossRef]
- Viereck, L.A. The Alaska Vegetation Classification; General technical report; USDA Pacific Northwest Research Station: Corvallis, OR, USA, 1992.
- Narita, K.; Harada, K.; Saito, K.; Sawada, Y.; Fukuda, M.; Tsuyuzaki, S. Vegetation and Permafrost Thaw Depth 10 Years after a Tundra Fire in 2002, Seward Peninsula, Alaska. Arct. Antarct. Alp. Res. 2015, 47, 547–559. [Google Scholar] [CrossRef] [Green Version]
- Hinzman, L.D.; Kane, D.L.; Yoshikawa, K.; Carr, A.; Bolton, W.R.; Fraver, M. Hydrological variations among watersheds with varying degrees of permafrost. In Proceedings of the 8th International Conference on Permafrost, Zurich, Switzerland, 21–25 July 2003; A.A. Balkema: Exton, PA, USA, 2003; pp. 407–411. [Google Scholar]
- Walker, D.A.; Breen, A.L.; Druckenmiller, L.A.; Wirth, L.W.; Fisher, W.; Raynolds, M.K.; Sibík, J.; Walker, M.D.; Hennekens, S.; Boggs, K.; et al. The Alaska Arctic Vegetation Archive (AVA-AK). Phytocoenologia 2016, 46, 221–229. [Google Scholar] [CrossRef] [Green Version]
- Raynolds, M.K.; Walker, D.A.; Maier, H.A. Plant community-level mapping of arctic Alaska based on the Circumpolar Arctic Vegetation Map. Phytocoenologia 2005, 35, 821–848. [Google Scholar] [CrossRef]
- Hartigan, J.A. Clustering Algorithms; John Wiley & Sons: Hoboken, NJ, USA, 1975; p. 351. [Google Scholar]
- Hoffman, F.M.; Hargrove, W.W.; Mills, R.T.; Mahajan, S.; Erickson, D.J.; Oglesby, R.J. Multivariate Spatio-Temporal Clustering (MSTC) as a Data Mining Tool for Environmental Applications. In Proceedings of the iEMSs Fourth Biennial Meeting: International Congress on Environmental Modelling and Software Society (iEMSs 2008), Barcelona, Spain, 7–10 July 2008; pp. 1774–1781. [Google Scholar]
- Bradley, P.S.; Fayyad, U.M. Refining Initial Points for K-Means Clustering. In Proceedings of the Fifteenth International Conference on Machine Learning, Madison, WI, USA, 24–27 July 1998; Morgan Kaufmann Publishers Inc.: San Francisco, CA, USA, 1998; pp. 91–99. [Google Scholar]
- Kumar, J.; Mills, R.T.; Hoffman, F.M.; Hargrove, W.W. Parallel k-Means Clustering for Quantitative Ecoregion Delineation Using Large Data Sets. In Proceedings of the International Conference on Computational Science (ICCS 2011), Singapore, 1–3 June 2011; Sato, M., Matsuoka, S., Sloot, P.M., van Albada, G.D., Dongarra, J., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; Volume 4, pp. 1602–1611. [Google Scholar] [CrossRef]
- Halko, N.; Martinsson, P.G.; Tropp, J.A. Finding structure with randomness: Probabilistic algorithms for constructing approximate matrix decompositions. arXiv, 2009; arXiv:math.NA/0909.4061. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Advances in Neural Information Processing Systems 25; Pereira, F., Burges, C.J.C., Bottou, L., Weinberger, K.Q., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2012; pp. 1097–1105. [Google Scholar]
- Zeiler, M.D.; Fergus, R. Visualizing and Understanding Convolutional Networks. arXiv, 2013; arXiv:1311.2901. [Google Scholar]
- Springenberg, J.T.; Dosovitskiy, A.; Brox, T.; Riedmiller, M.A. Striving for Simplicity: The All Convolutional Ne. arXiv, 2014; arXiv:1412.6806. [Google Scholar]
- Chollet, F. Keras. 2015. Available online: https://github.com/fchollet/keras (accessed on 1 February 2018).
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. TensorFlow: A system for large-scale machine learning. In Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI 16), Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Wang, T.; Zhang, H.; Lin, H.; Fang, C. Textural–Spectral Feature-Based Species Classification of Mangroves in Mai Po Nature Reserve from Worldview-3 Imagery. Remote Sens. 2016, 8, 24. [Google Scholar] [CrossRef]
- Hinkel, K.M.; Nelson, F.E. Summer Differences among Arctic Ecosystems in Regional Climate Forcing. J. Clim. 2000, 13, 2002–2010. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Nets. In Advances in Neural Information Processing Systems 27; Ghahramani, Z., Welling, M., Cortes, C., Lawrence, N.D., Weinberger, K.Q., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2014; pp. 2672–2680. [Google Scholar]
- Shrivastava, A.; Pfister, T.; Tuzel, O.; Susskind, J.; Wang, W.; Webb, R. Learning from Simulated and Unsupervised Images through Adversarial Training. arXiv, 2016; arXiv:1612.07828. [Google Scholar]
- Fu, G.; Liu, C.; Zhou, R.; Sun, T.; Zhang, Q. Classification for High Resolution Remote Sensing Imagery Using a Fully Convolutional Network. Remote Sens. 2017, 9, 498. [Google Scholar] [CrossRef]
- Jindal, I.; Nokleby, M.S.; Chen, X. Learning Deep Networks from Noisy Labels with Dropout Regularization. arXiv, 2017; arXiv:1705.03419. [Google Scholar]
- Wang, S.; Liu, W.; Wu, J.; Cao, L.; Meng, Q.; Kennedy, P.J. Training deep neural networks on imbalanced data sets. In Proceedings of the 2016 International Joint Conference on Neural Networks (IJCNN), Vancouver, BC, Canada, 24–29 July 2016; pp. 4368–4374. [Google Scholar] [CrossRef]
- Yang, J.; Zhao, Y.Q.; Chan, J.C.W. Hyperspectral and Multispectral Image Fusion via Deep Two-Branches Convolutional Neural Network. Remote Sens. 2018, 10, 800. [Google Scholar] [CrossRef]
- Panboonyuen, T.; Jitkajornwanich, K.; Lawawirojwong, S.; Srestasathiern, P.; Vateekul, P. Road Segmentation of Remotely-Sensed Images Using Deep Convolutional Neural Networks with Landscape Metrics and Conditional Random Fields. Remote Sens. 2017, 9, 680. [Google Scholar] [CrossRef]
- Langford, Z.; Kumar, J.; Hoffman, F.; Iversen, C.; Breen, A. Remote Sensing-Based, 5-m, Vegetation Distributions, Kougarok Study Site, Seward Peninsula, Alaska, ca. 2000–2016; Oak Ridge National Laboratory (ORNL): Oak Ridge, TN, USA, 2018. [CrossRef]













| AKEVT Class | SR Area | KW Area | AATVM Class | Samples | Plot Size |
|---|---|---|---|---|---|
| Alder-Willow Shrub | 72.38 | 0.57 | Alder Shrubland | 5 | 5 × 5 m |
| Willow-Birch Tundra | 5 | 2.5 × 2.5 m | |||
| Mixed Shrub-Sedge Tussock Tundra-Bog | 117.61 | 0.45 | Tussock Tundra | 5 | 2.5 × 2.5 m |
| Shrubby Tussock Tundra | 5 | 2.5 × 2.5 m | |||
| Dryas/Lichen Dwarf Shrub Tundra | 20.02 | 0.15 | Dwarf Shrub Lichen Tundra | 5 | 2.5 × 2.5 m |
| Non-acidic Mountain Complex | 5 | 2.5 × 2.5 m | |||
| Sedge-Willow-Dryas Tundra | 113.34 | 0.54 | |||
| Non-Vegetated | 5.53 | 0.007 | |||
| Water | 6.00 | 0.02 |
| Sensor Group | Predictor Variable | Unit | Collection Date | Resolution |
|---|---|---|---|---|
| SPOT-5 | Green, Red, NIR (0.5–0.9 μm) | DN | June–September 2009–2012 | 2.5 m |
| DSM | Elevation | meter | July 2012 | 5 m |
| EO-1 | 198 spectral bands (0.4–2.5 μm) | DN | 24 June 2015 | 30 m |
| Landsat 8 | 9 spectral bands (0.4–2.29 μm) | DN | 17 August 2016 | 30 m |
| AKEVT Land Cover | Study Region Area (km2) | Kougarok Watershed Area (km2) |
|---|---|---|
| Alder-Willow Shrub | 73.33 | 0.55 |
| Mixed Shrub-Sedge Tussock Tundra-Bog | 135.27 | 0.58 |
| Dryas/Lichen Dwarf Shrub Tundra | 14.88 | 0.07 |
| Sedge-Willow-Dryas Tundra | 102.14 | 0.50 |
| Non-Vegetated | 2.24 | 0.02 |
| Water | 7.04 | 0.04 |
| Total | 334.89 | 1.75 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Langford, Z.L.; Kumar, J.; Hoffman, F.M.; Breen, A.L.; Iversen, C.M. Arctic Vegetation Mapping Using Unsupervised Training Datasets and Convolutional Neural Networks. Remote Sens. 2019, 11, 69. https://doi.org/10.3390/rs11010069
Langford ZL, Kumar J, Hoffman FM, Breen AL, Iversen CM. Arctic Vegetation Mapping Using Unsupervised Training Datasets and Convolutional Neural Networks. Remote Sensing. 2019; 11(1):69. https://doi.org/10.3390/rs11010069
Chicago/Turabian StyleLangford, Zachary L., Jitendra Kumar, Forrest M. Hoffman, Amy L. Breen, and Colleen M. Iversen. 2019. "Arctic Vegetation Mapping Using Unsupervised Training Datasets and Convolutional Neural Networks" Remote Sensing 11, no. 1: 69. https://doi.org/10.3390/rs11010069