Supervised Classification of RGB Aerial Imagery to Evaluate the Impact of a Root Rot Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Data Collection and Processing
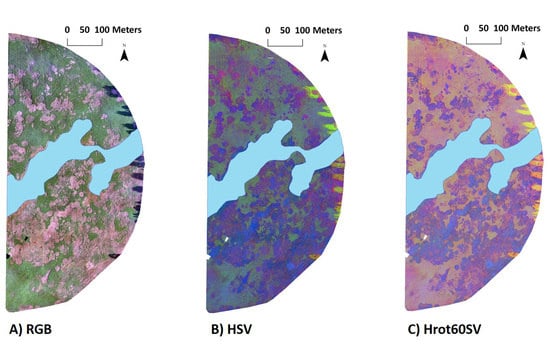
2.3. Image Classification
2.4. Model Accuracy
2.5. Agreement between Two Classified Images
2.6. Post-Processing of Classified Images
3. Results
3.1. UAS-Acquired Images
3.2. Manned-Aircraft-Acquired Images
3.3. Effects of Post-Processing on Image Accuracy
3.4. Effect of PRR Disease on Alfalfa Stand
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Dash, J.P.; Watt, M.S.; Pearse, G.D.; Heaphy, M.; Dungey, H.S. Assessing very high resolution UAV imagery for monitoring forest health during a simulated disease outbreak. ISPRS J. Photogramm. Remote Sens. 2017, 131, 1–14. [Google Scholar] [CrossRef]
- Mahlein, A.-K. Plant disease detection by imaging sensors–parallels and specific demands for precision agriculture and plant phenotyping. Plant Dis. 2016, 100, 241–251. [Google Scholar] [CrossRef]
- Simko, I.; Jimenez-Berni, J.A.; Sirault, X.R. Phenomic approaches and tools for phytopathologists. Phytopathology 2017, 107, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.L.; Di Bello, P.L.; Lear, M.; Balci, Y. Bacterial leaf scorch in the District of Columbia: Distribution, host range, and presence of Xylella fastidiosa among urban trees. Plant Dis. 2014, 98, 1611–1618. [Google Scholar] [CrossRef]
- Mirik, M.; Jones, D.C.; Price, J.A.; Workneh, F.; Ansley, R.J.; Rush, C.M. Satellite remote sensing of wheat infected by Wheat streak mosaic virus. Plant Dis. 2011, 95, 4–12. [Google Scholar] [CrossRef]
- USDA-NASS. Available online: http://quickstats.nass.usda.gov/results/7C288133-3D6D-356D-92EB-E0D2FDB11C76 (accessed on 20 April 2018).
- Taubenhaus, J.J.; Ezekiel, W.N. A rating of plants with reference to their relative resistance or susceptibility to Phymatotrichum root rot. Texas Agric. Exp. Stn Bull. 1936, 527, 1–52. [Google Scholar]
- Streets, R.B.; Bloss, H.E. Phymatotrichum Root Rot; American Phytopathological Society: St. Paul, MN, USA, 1973; Volume Monograph No. 8. [Google Scholar]
- Young, C.A.; Uppalapati, S.R.; Mysore, K.S.; Marek, S.M. Phymatotrichopsis root rot. In Compendium of Alfalfa Diseases and Pests, 3rd ed.; Samac, D.A., Rhodes, L.H., Lamp, W.O., Eds.; American Phytopathological Society: St. Paul, MN, USA, 2015; pp. 44–46. [Google Scholar]
- King, C.J. Habits of the cotton root rot fungus. J. Agric. Res. 1923, 26, 405–418. [Google Scholar]
- Steddom, K.; Jones, D.; Rush, C. A picture is worth a thousand words. APSnet Features 2005. [Google Scholar] [CrossRef]
- Taubenhaus, J.J.; Ezekiel, W.N.; Neblette, C.B. Airplane photography in the study of cotton root rot. Phytopathology 1929, 19, 1025–1029. [Google Scholar]
- Nixon, P.R.; Escobar, D.E.; Bowen, R.L. A multispectral false-color video imaging system for remote sensing applications. In Proceedings of the 11th Biennial Workshop on Color Aerial Photography and Videography in the Plant Sciences and Related Fields, Weslaco, TX, USA, 27 April–1 May 1987; pp. 295–305. [Google Scholar]
- Nixon, P.R.; Lyda, S.D.; Heilman, M.D.; Bowen, R.L.; Texas A&M Agricultural Experiment Station, College Station. Incidence and Control of Cotton Root Rot Observed with Color Infrared Photography; Information Systems Division, National Agricultural Library: Beltsville, MD, USA, 1975. [Google Scholar]
- Yang, C.; Everitt, J.H.; Fernandez, C.J. Comparison of airborne multispectral and hyperspectral imagery for mapping cotton root rot. Biosyst. Eng. 2010, 107, 131–139. [Google Scholar] [CrossRef]
- Yang, C.; Odvody, G.N.; Fernandez, C.J.; Landivar, J.A.; Minzenmayer, R.R.; Nichols, R.L. Evaluating unsupervised and supervised image classification methods for mapping cotton root rot. Precis. Agric. 2015, 16, 201–215. [Google Scholar] [CrossRef]
- Yang, C.; Odvody, G.N.; Thomasson, J.A.; Isakeit, T.; Nichols, R.L. Change detection of cotton root rot infection over 10-year intervals using airborne multispectral imagery. Comput. Electron. Agric. 2016, 123, 154–162. [Google Scholar] [CrossRef] [Green Version]
- Aylor, D.E.; Schmale, D.G., III; Shields, E.J.; Newcomb, M.; Nappo, C.J. Tracking the potato late blight pathogen in the atmosphere using unmanned aerial vehicles and Lagrangian modeling. Agric. For. Meteorol. 2011, 151, 251–260. [Google Scholar] [CrossRef]
- Mattupalli, C.; Komp, M.R.; Young, C.A. Integrating geospatial technologies and unmanned aircraft systems into the grower’s disease management toolbox. APS Features 2017. [Google Scholar] [CrossRef]
- Sankaran, S.; Khot, L.R.; Espinoza, C.Z.; Jarolmasjed, S.; Sathuvalli, V.R.; Vandemark, G.J.; Miklas, P.N.; Carter, A.H.; Pumphrey, M.O.; Knowles, N.R.; et al. Low-altitude, high-resolution aerial imaging systems for row and field crop phenotyping: A review. Eur. J. Agron. 2015, 70, 112–123. [Google Scholar] [CrossRef]
- Zhang, C.; Kovacs, J.M. The application of small unmanned aerial systems for precision agriculture: A review. Precis. Agric. 2012, 13, 693–712. [Google Scholar] [CrossRef]
- Calderón, R.; Montes-Borrego, M.; Landa, B.B.; Navas-Cortés, J.A.; Zarco-Tejada, P.J. Detection of downy mildew of opium poppy using high-resolution multi-spectral and thermal imagery acquired with an unmanned aerial vehicle. Precis. Agric. 2014, 15, 639–661. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Ruiz, F.; Sankaran, S.; Maja, J.M.; Lee, W.S.; Rasmussen, J.; Ehsani, R. Comparison of two aerial imaging platforms for identification of Huanglongbing-infected citrus trees. Comput. Electron. Agric. 2013, 91, 106–115. [Google Scholar] [CrossRef]
- Booth, D.T.; Cox, S.E. Very large scale aerial photography for rangeland monitoring. Geocarto Int. 2006, 21, 27–34. [Google Scholar] [CrossRef]
- Booth, D.T.; Cox, S.E.; Berryman, R.D. Precision measurements from very-large scale aerial digital imagery. Environ. Monit. Assess. 2006, 112, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Moffet, C.A.; Taylor, J.B.; Booth, D.T. Very large scale aerial (VLSA) imagery for assessing postfire bitterbrush recovery. In Proceedings of the USDA Forest Service RMRS-P-52, Cedar City, UT, USA, 6–8 June 2006; pp. 161–168. [Google Scholar]
- Hijmans, R.J.; van Etten, J.; Cheng, J.; Mattiuzzi, M.; Sumner, M.; Greenberg, J.A.; Lamigueiro, O.P.; Bevan, A.; Racine, E.B.; Shortridge, A.; et al. Raster: Geographic data analysis and modeling. In R Package Version 2.6-7; R Core Team: Vienna, Austria, 2017. [Google Scholar]
- Kuhn, M.; Wing, J.; Weston, S.; Williams, A.; Keefer, C.; Engelhardt, A.; Cooper, T.; Mayer, Z.; Kenkel, B.; The R Core Team; et al. Caret: Classification and regression training. In R Package Version 6.0-78; R Core Team: Vienna, Austria, 2017. [Google Scholar]
- Yang, C.; Odvody, G.N.; Fernandez, C.J.; Landivar, J.A.; Minzenmayer, R.R.; Nichols, R.L.; Thomasson, J.A. Monitoring cotton root rot progression within a growing season using airborne multispectral imagery. J. Cotton Sci. 2014, 18, 85–93. [Google Scholar]
- An, N.; Palmer, C.M.; Baker, R.L.; Markelz, R.C.; Ta, J.; Covington, M.F.; Maloof, J.N.; Welch, S.M.; Weinig, C. Plant high-throughput phenotyping using photogrammetry and imaging techniques to measure leaf length and rosette area. Comput. Electron. Agric. 2016, 127, 376–394. [Google Scholar] [CrossRef] [Green Version]
- Třebický, V.; Fialová, J.; Kleisner, K.; Havlíček, J. Focal length affects depicted shape and perception of facial images. PLoS ONE 2016, 11, e0149313. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Hernández, J.L.; García-Mateos, G.; González-Esquiva, J.M.; Escarabajal-Henarejos, D.; Ruiz-Canales, A.; Molina-Martínez, J.M. Optimal color space selection method for plant/soil segmentation in agriculture. Comput. Electron. Agric. 2016, 122, 124–132. [Google Scholar] [CrossRef]
- Philipp, I.; Rath, T. Improving plant discrimination in image processing by use of different colour space transformations. Comput. Electron. Agric. 2002, 35, 1–15. [Google Scholar] [CrossRef]
- García-Mateos, G.; Hernández-Hernández, J.L.; Escarabajal-Henarejos, D.; Jaen-Terrones, S.; Molina-Martínez, J.M. Study and comparison of color models for automatic image analysis in irrigation management applications. Agric. Water Manag. 2015, 151, 158–166. [Google Scholar] [CrossRef]
- Chaves-González, J.M.; Vega-Rodríguez, M.A.; Gómez-Pulido, J.A.; Sánchez-Pérez, J.M. Detecting skin in face recognition systems: A colour spaces study. Digit. Signal Process. 2010, 20, 806–823. [Google Scholar] [CrossRef]
- Hamuda, E.; Mc Ginley, B.; Glavin, M.; Jones, E. Automatic crop detection under field conditions using the HSV colour space and morphological operations. Comput. Electron. Agric. 2017, 133, 97–107. [Google Scholar] [CrossRef]
- Huang, W.; Kim, K.; Yang, Y.; Kim, Y.S. Automatic shadow removal by illuminance in HSV color space. Comput. Sci. Inf. Technol. 2015, 3, 70–75. [Google Scholar] [CrossRef]
- Petropoulos, G.P.; Vadrevu, K.P.; Xanthopoulos, G.; Karantounias, G.; Scholze, M. A comparison of spectral angle mapper and artificial neural network classifiers combined with Landsat TM imagery analysis for obtaining burnt area mapping. Sensors 2010, 10, 1967–1985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamuda, E.; Glavin, M.; Jones, E. A survey of image processing techniques for plant extraction and segmentation in the field. Comput. Electron. Agric. 2016, 125, 184–199. [Google Scholar] [CrossRef]
- Jeon, H.Y.; Tian, L.F.; Zhu, H. Robust crop and weed segmentation under uncontrolled outdoor illumination. Sensors 2011, 11, 6270–6283. [Google Scholar] [CrossRef] [PubMed]





| Platform | Image (Acquisition Date) | Ground Sampling Distance (m) | Flight Time (Hours, Central Standard Time) |
|---|---|---|---|
| UAS | June 2014 (23 June 2014) | 0.018 | 13:23 to 14:13 |
| UAS | August 2014 (5 August 2014) | 0.024 | 11:49 to 12:35 |
| Manned aircraft | October 2014 (29 October 2014) | 0.046 | 10:23 to 11:35 |
| Manned aircraft | June 2015 (29 June 2015) | 0.040 | 13:23 to 14:13 |
| Manned aircraft | September 2015 (17 September 2015) | 0.064 | 8:02 to 9:00 |
| Manned aircraft | October 2015 (15 October 2015) | 0.063 | 8:37 to 9:22 |
| Image | Training Pixels | Validation Pixels | ||||
|---|---|---|---|---|---|---|
| Alfalfa | Soil | Weed | Alfalfa | Soil | Weed | |
| June 2014 | 1865 | 2057 | 204 | 1241 | 1406 | 192 |
| August 2014 | 1587 | 1667 | 569 | 898 | 957 | 591 |
| October 2014 | 1428 | 1505 | 568 | 822 | 819 | 513 |
| June 2015 | 984 | 1029 | 502 | 613 | 636 | 577 |
| September 2015 | 1436 | 1529 | 472 | 987 | 956 | 576 |
| October 2015 | 1515 | 1412 | 356 | 888 | 893 | 416 |
| Image | Image Acquired Platform | Spectral Signature Y | Balanced Accuracy Z | Overall Mean Accuracy Z | ||
|---|---|---|---|---|---|---|
| Alfalfa | Soil | Weed | ||||
| June 2014 | UAS | Image specific | 0.968 ± 0.02 | 0.999 ± 0 | 0.899 ± 0.08 | 0.968 ± 0.04 |
| UAS platform specific | 0.9 ± 0.03 | 0.998 ± 0.01 | 0.841 ± 0.10 | 0.901 ± 0.03 | ||
| Manned aircraft platform specific | 0.5 ± 0 | 0.971 ± 0.02 | 0.736 ± 0.03 | 0.508 ± 0.01 | ||
| August 2014 W | UAS | UAS platform specific | 0.871 ± 0.05 | 1 ± 0 | 0.905 ± 0.04 | 0.896 ± 0.04 |
| Manned aircraft platform specific | 0.5 ± 0 | 0.864 ± 0.03 | 0.770 ± 0.04 | 0.584 ± 0.03 | ||
| October 2014 | Manned aircraft | Image specific | 0.986 ± 0.02 | 0.962 ± 0.03 | 0.967 ± 0.03 | 0.959 ± 0.03 |
| UAS platform specific | 0.731 ± 0.06 | 0.659 ± 0.04 | 0.509 ± 0.02 | 0.560 ± 0.05 | ||
| Manned aircraft platform specific | 0.684 ± 0.06 | 0.934 ± 0.03 | 0.686 ± 0.07 | 0.687 ± 0.06 | ||
| June 2015 | Manned aircraft | Image specific | 0.976 ± 0.02 | 1 ± 0 | 0.983 ± 0.02 | 0.981 ± 0.02 |
| UAS platform specific | 0.559 ± 0.06 | 0.777 ± 0.04 | 0.507 ± 0.02 | 0.535 ± 0.04 | ||
| Manned aircraft platform specific | 0.718 ± 0.05 | 0.988 ± 0.02 | 0.846 ± 0.04 | 0.781 ± 0.04 | ||
| September 2015 X | Manned aircraft | UAS platform specific | 0.516 ± 0.04 | 0.547 ± 0.03 | 0.5 ± 0 | 0.405 ± 0.02 |
| Image specific | 0.928 ± 0.04 | 0.978 ± 0.02 | 0.887 ± 0.06 | 0.915 ± 0.04 | ||
| October 2015 | Manned aircraft | Image specific | 0.954 ± 0.03 | 0.981 ± 0.02 | 0.897 ± 0.06 | 0.940 ± 0.03 |
| UAS platform specific | 0.564 ± 0.04 | 0.543 ± 0.03 | 0.5 ± 0 | 0.457 ± 0.03 | ||
| Manned aircraft platform specific | 0.946 ± 0.04 | 0.956 ± 0.03 | 0.812 ± 0.08 | 0.903 ± 0.04 | ||
| Class Pair (Image Specific–UAS Platform Specific) | Percentage of Pixels | Weight Factor Z |
|---|---|---|
| Alfalfa–Alfalfa | 57.65 | - |
| Soil–Soil | 17.92 | - |
| Weed–Weed | 0.70 | - |
| Alfalfa–Soil | 3.08 | 0.130 |
| Alfalfa–Weed | 7.38 | 0.311 |
| Soil–Alfalfa | 0.03 | 0.001 |
| Soil–Weed | 0.01 | 0.001 |
| Weed–Alfalfa | 6.77 | 0.285 |
| Weed–Soil | 6.46 | 0.272 |
| Class Pair (Image Specific–UAS Platform Specific) | Weight Factor X | Manual Classification (Percentage of Pixels) Y | Image Specific Correct Z | UAS Platform Specific correct Z | Neither Correct Z | ||
|---|---|---|---|---|---|---|---|
| Alfalfa | Soil | Weed | |||||
| Alfalfa–Soil | 0.130 | 45 | 55 | 0 | 5.85 | 7.15 | 0.00 |
| Alfalfa–Weed | 0.311 | 100 | 0 | 0 | 31.10 | 0.00 | 0.00 |
| Soil–Alfalfa | 0.001 | 70 | 30 | 0 | 0.03 | 0.07 | 0.00 |
| Soil–Weed | 0.001 | 60 | 40 | 0 | 0.04 | 0.00 | 0.06 |
| Weed–Alfalfa | 0.285 | 80 | 20 | 0 | 0.00 | 22.80 | 5.70 |
| Weed–Soil | 0.272 | 20 | 65 | 15 | 4.08 | 17.68 | 5.44 |
| 41.10 | 47.70 | 11.20 | |||||
| Image | Image Acquisition Platform/Spectral Signature | Balanced Accuracy Z | Overall Mean Accuracy Z | ||
|---|---|---|---|---|---|
| Alfalfa | Soil | Weed | |||
| June 2014 | UAS/UAS platform specific | 0.897 ± 0 | 0.998 ± 0.01 | 0.78 ± 0.04 | 0.898 ± 0.01 |
| August 2014 | UAS/UAS platform specific | 0.951 ± 0.03 | 1 ± 0 | 0.975 ± 0.01 | 0.963 ± 0.02 |
| October 2014 | Manned aircraft/Image specific | 1 ± 0 | 0.951 ± 0.03 | 0.935 ± 0.01 | 0.953 ± 0.02 |
| June 2015 | Manned aircraft/Image specific | 1 ± 0 | 1 ± 0 | 1 ± 0 | 1 ± 0 |
| September 2015 | Manned aircraft/Image specific | 1 ± 0 | 1 ± 0 | 1 ± 0 | 1 ± 0 |
| October 2015 | Manned aircraft/Image specific | 1 ± 0 | 0.998 ± 0.01 | 0.994 ± 0.02 | 0.998 ± 0.01 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mattupalli, C.; Moffet, C.A.; Shah, K.N.; Young, C.A. Supervised Classification of RGB Aerial Imagery to Evaluate the Impact of a Root Rot Disease. Remote Sens. 2018, 10, 917. https://doi.org/10.3390/rs10060917
Mattupalli C, Moffet CA, Shah KN, Young CA. Supervised Classification of RGB Aerial Imagery to Evaluate the Impact of a Root Rot Disease. Remote Sensing. 2018; 10(6):917. https://doi.org/10.3390/rs10060917
Chicago/Turabian StyleMattupalli, Chakradhar, Corey A. Moffet, Kushendra N. Shah, and Carolyn A. Young. 2018. "Supervised Classification of RGB Aerial Imagery to Evaluate the Impact of a Root Rot Disease" Remote Sensing 10, no. 6: 917. https://doi.org/10.3390/rs10060917
APA StyleMattupalli, C., Moffet, C. A., Shah, K. N., & Young, C. A. (2018). Supervised Classification of RGB Aerial Imagery to Evaluate the Impact of a Root Rot Disease. Remote Sensing, 10(6), 917. https://doi.org/10.3390/rs10060917