Hyperspectral Imagery Classification Based on Semi-Supervised Broad Learning System
Abstract
:1. Introduction
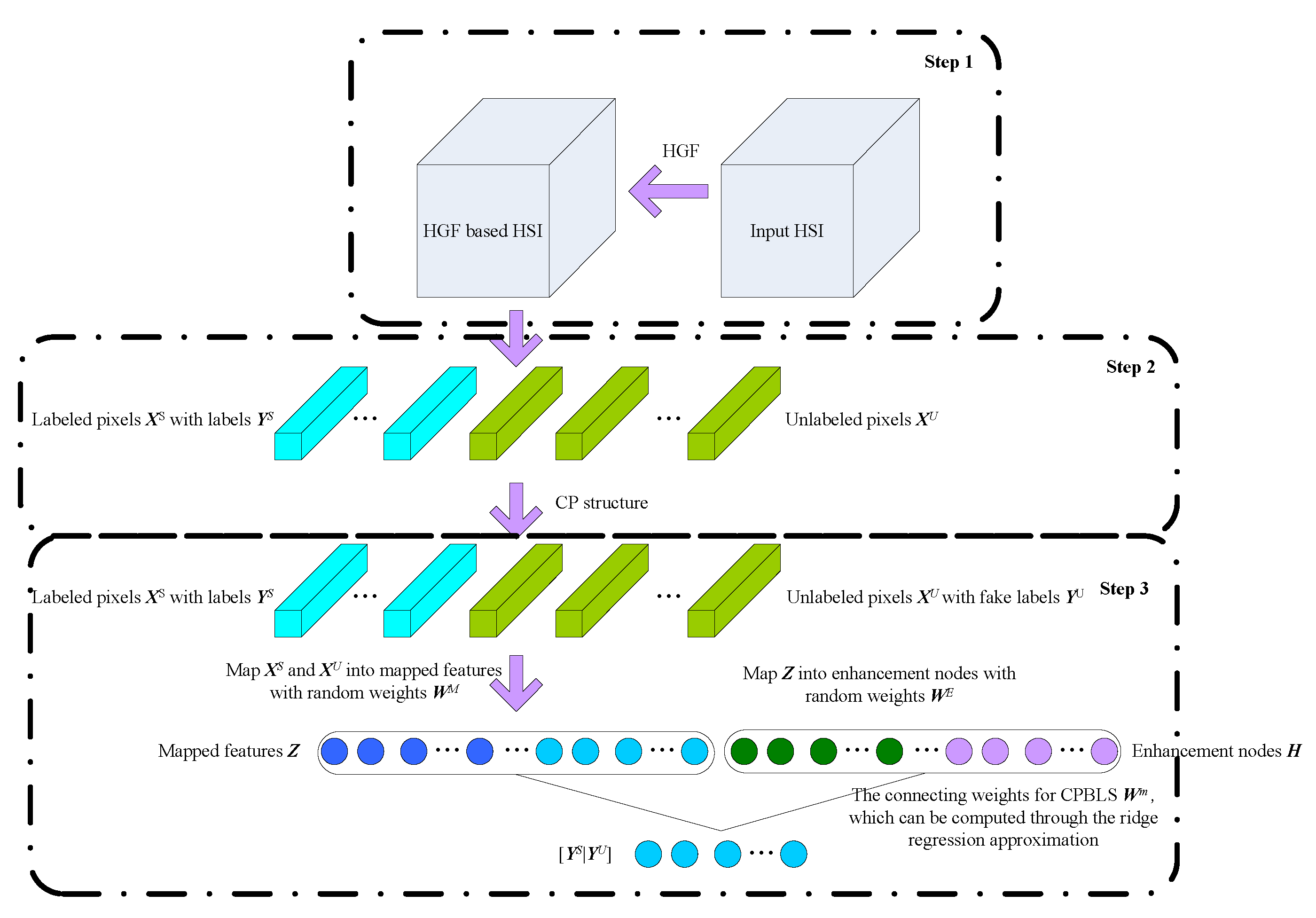
2. HSI Classification Based On SBLS
2.1. Hierarchical Guidance Filtering
2.2. Class-Probability Structure
2.3. SBLS
3. Experiments and Analysis
3.1. HSI Datasets
3.2. Comparative Experiments
3.3. Parameter Analysis
4. Discussion
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Gao, L.; Yang, B.; Du, Q.; Zhang, B. Adjusted spectral matched filter for target detection in hyperspectral imagery. Remote Sens. 2015, 7, 6611–6634. [Google Scholar] [CrossRef]
- Onoyama, H.; Ryu, C.; Suguri, M.; Lida, M. Integrate growing temperature to estimate the nitrogen content of rice plants at the heading stage using hyperspectral imagery. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2506–2515. [Google Scholar] [CrossRef]
- Brunet, D.; Sills, D. A generalized distance transform: Theory and applications to weather analysis and forecasting. IEEE Trans. Geosci. Remote Sens. 2017, 55, 1752–1764. [Google Scholar] [CrossRef]
- Islam, T.; Hulley, G.C.; Malakar, N.K.; Radocinski, R.G.; Guillevic, P.C.; Hook, S.J. A physics-based algorithm for the simultaneous retrieval of land surface temperature and emissivity from VIIRS thermal infrared data. IEEE Trans. Geosci. Remote Sens. 2017, 55, 563–576. [Google Scholar] [CrossRef]
- Li, W.; Du, Q.; Zhang, F.; Hu, W. Collaborative-representation-based nearest neighbor classifier for hyperspectral imagery. IEEE Geosci. Remote Sens. Lett. 2015, 12, 389–393. [Google Scholar] [CrossRef]
- Wu, Y.F.; Yang, X.H.; Plaza, A.; Qiao, F.; Gao, L.R.; Zhang, B.; Cui, Y.B. Approximate computing of remotely Sensed data: SVM hyperspectral image classification as a case study. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 5806–5818. [Google Scholar] [CrossRef]
- Xue, Z.; Du, P.; Su, H. Harmonic analysis for hyperspectral image classification integrated with PSO optimized SVM. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2131–2146. [Google Scholar] [CrossRef]
- Li, W.; Chen, C.; Su, H.; Du, Q. Local binary patterns and extreme learning machine for hyperspectral imagery classification. IEEE Trans. Geosci. Remote Sens. 2015, 53, 3681–3693. [Google Scholar] [CrossRef]
- Alom, M.; Sidike, P.; Taha, T.; Asari, V. State preserving extreme learning machine: A monotonically increasing learning approach. Neural Process. Lett. 2016, 45, 703–725. [Google Scholar] [CrossRef]
- Feng, S.; Chen, C.L.P. A fuzzy restricted boltzmann machine: Novel learning algorithms based on crisp possibilistic mean value of fuzzy numbers. IEEE Trans. Fuzzy Syst. 2016. [Google Scholar] [CrossRef]
- Chen, C.L.P.; Zhang, C.Y.; Chen, L.; Gan, M. Fuzzy restricted boltzmann machine for the enhancement of deep learning. IEEE Trans. Fuzzy Syst. 2015, 23, 2163–2173. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, Z.; Zhao, X.; Wang, G. Deep learning-based classification of hyperspectral data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2094–2107. [Google Scholar] [CrossRef]
- Tao, C.; Pan, H.; Li, Y.; Zou, Z. Unsupervised spectral-spatial feature learning with stacked sparse autoencoder for hyperspectral imagery classification. IEEE Geosci. Remote Sens. Lett. 2015, 12, 2438–2442. [Google Scholar]
- Chen, Y.; Zhao, X.; Jia, X. Spectral-spatial classification of hyperspectral data based on deep belief network. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 1–12. [Google Scholar] [CrossRef]
- Romero, A.; Gatta, C.; Camps-Valls, G. Unsupervised deep feature extraction for remote sensing image classification. IEEE Trans. Geosci. Remote Sens. 2016, 54, 1349–1362. [Google Scholar] [CrossRef]
- Li, W.; Wu, G.; Zhang, F.; Du, Q. Hyperspectral image classification using deep pixel-pair features. IEEE Trans. Geosci. Remote Sens. 2017, 55, 844–853. [Google Scholar] [CrossRef]
- Santara, A.; Mani, K.; Hatwar, P.; Singh, A.; Garg, A.; Padia, K.; Mitra, P. BASS net: Band-adaptive spectral-spatial feature learning neural network for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2017. [Google Scholar] [CrossRef]
- Pan, B.; Shi, Z.; Zhang, N.; Xie, S. Hyperspectral image classification based on nonlinear spectral-spatial network. IEEE Geosci. Remote Sens. Lett. 2016, 13, 1782–1786. [Google Scholar] [CrossRef]
- Pan, B.; Shi, Z.; Xu, X. R-VCANet: A new deep-learning-based hyperspectral image classification method. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10, 1975–1986. [Google Scholar] [CrossRef]
- Chen, C.L.P.; Liu, Z. Broad learning system: An effective and efficient incremental learning system without the need for deep architecture. IEEE Trans. Neural Netw. Learn. Syst. 2017. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.L.P.; Wan, J.Z. A rapid learning and dynamic stepwise updating algorithm for flat neural networks and the application to time-series prediction. IEEE Trans. Syst. Man Cybern. Part B 1999, 29, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.L.P. A rapid supervised learning neural network for function interpolation and approximation. IEEE Trans. Neural Netw. 1996, 7, 1220–1230. [Google Scholar] [CrossRef] [PubMed]
- Camps-Valls, G.; Marsheva, T.B.; Zhou, D. Semi-supervised graph-based hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2007, 45, 3044–3054. [Google Scholar] [CrossRef]
- Belkin, M.; Niyogi, P.; Sindhwani, V. Manifold regularization: Ageometric framework for learning from examples. J. Mach. Learn. Res. 2006, 7, 2399–2434. [Google Scholar]
- Wang, F.; Zhang, C. Label propagation through linear neighborhoods. IEEE Trans. Knowl. Data Eng. 2008, 20, 55–67. [Google Scholar] [CrossRef]
- Zhuang, L.; Gao, S.; Tang, J.; Wang, J.; Lin, Z.; Ma, Y.; Yu, N. Constructing a nonnegative low-rank and sparse graph with data-adaptive features. IEEE Trans. Image Process. 2015, 24, 3717–3728. [Google Scholar] [CrossRef] [PubMed]
- De Morsier, F.; Borgeaud, M.; Gass, V.; Thiran, J.P.; Tuia, D. Kernel low-rank and sparse graph for unsupervised and semi-supervised classification of hyperspectral images. IEEE Trans. Geosci. Remote Sens. 2016, 54, 3410–3420. [Google Scholar] [CrossRef]
- Shao, Y.J.; Sang, N.; Gao, C.X.; Ma, L. Probabilistic class structure regularized sparse representation graph for semi-supervised hyperspectral image classification. Pattern Recognit. 2017, 63, 102–114. [Google Scholar] [CrossRef]
- Pan, B.; Shi, Z.W.; Xu, X. Hierarchical guidance filtering based ensemble classification for hyperspectral image. IEEE Trans. Geosci. Remote Sens. 2017, 55, 4177–4189. [Google Scholar] [CrossRef]





| Input: HGF-based HSI spectral-spatial representation. (a) Calculate class-probability matrix according to Equation (5). (b) Calculate pseudo labels for unlabeled samples according to Equation (6). (c) Calculate and according to Equations (7)–(8), respectively. (d) Calculate weights of BLS according to Equations (12)–(13). (e) Calculate predictive labels with Equations (7), (8), and (14), according to , , , , and . Output: predictive labels . |
| No. | Indian Pines | Salinas | Botswana | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Surface Object | s.l.s. | s.u.s. | Surface Object | s.l.s. | s.u.s. | Surface Object | s.l.s. | s.u.s. | |
| 1 | Alfalfa | 20 | 26 | Brocoli_green_weeds_1 | 20 | 500 | Water | 20 | 250 |
| 2 | Corn-notill | 20 | 1408 | Brocoli_green_weeds_2 | 20 | 500 | Hippo grass | 20 | 81 |
| 3 | Corn-mintill | 20 | 810 | Fallow | 20 | 500 | Floodplain grasses1 | 20 | 231 |
| 4 | Corn | 20 | 217 | Fallow_rough_plow | 20 | 500 | Floodplain grasses2 | 20 | 195 |
| 5 | Grass-pasture | 20 | 463 | Fallow_smooth | 20 | 500 | Reeds1 | 20 | 249 |
| 6 | Grass-trees | 20 | 710 | Stubble | 20 | 500 | Riparian | 20 | 249 |
| 7 | Grass-pasture-mowed | 20 | 8 | Celery | 20 | 500 | Firescar2 | 20 | 239 |
| 8 | Hay-windrowed | 20 | 458 | Grapes_untrained | 20 | 500 | Island interior | 20 | 183 |
| 9 | Oats | 10 | 10 | Soil_vinyard_develop | 20 | 500 | Acacia woodlands | 20 | 294 |
| 10 | Soybean-notill | 20 | 952 | Corn_senesced_green_weeds | 20 | 500 | Acacia shrublands | 20 | 228 |
| 11 | Soybean-mintill | 20 | 2435 | Lettuce_romaine_4wk | 20 | 500 | Acacia grasslands | 20 | 285 |
| 12 | Soybean-clean | 20 | 573 | Lettuce_romaine_5wk | 20 | 500 | Short mopane | 20 | 161 |
| 13 | Wheat | 20 | 185 | Lettuce_romaine_6wk | 20 | 500 | Mixed mopane | 20 | 248 |
| 14 | Woods | 20 | 1245 | Lettuce_romaine_7wk | 20 | 500 | Exposed soils | 20 | 75 |
| 15 | Buildings-Grass-Trees-Drives | 20 | 366 | Vinyard_untrained | 20 | 500 | |||
| 16 | Stone-Steel-Towers | 20 | 73 | Vinyard_vertical_trellis | 20 | 500 | |||
| Surface Object | SVM [6] | ELM [8] | SPELM [9] | SSG [23] | CNN-PPF [16] | BASS-Net [17] | R-VCANet [19] | HiFi-We [29] | BLS [20] | SBLS |
|---|---|---|---|---|---|---|---|---|---|---|
| Alfalfa (%) | 54.15 | 66.63 | 90.13 | 96.15 | 32.84 | 80.77 | 100 | 100 | 96.15 | 99.23 |
| Corn-notill (%) | 75.00 | 79.17 | 87.01 | 71.46 | 53.64 | 55.33 | 64.98 | 85.72 | 78.69 | 84.73 |
| Corn-mintill (%) | 72.11 | 87.93 | 85.30 | 84.07 | 56.36 | 59.75 | 86.05 | 91.36 | 98.52 | 94.49 |
| Corn (%) | 54.13 | 61.31 | 77.35 | 93.82 | 33.73 | 91.24 | 99.54 | 98.16 | 100 | 100 |
| Grass-pasture (%) | 87.88 | 98.58 | 94.31 | 85.40 | 79.34 | 89.42 | 91.14 | 91.14 | 96.33 | 90.67 |
| Grass-trees (%) | 98.15 | 99.40 | 99.62 | 97.35 | 94.67 | 94.93 | 99.30 | 99.86 | 90.14 | 99.86 |
| Grass-pasture-mowed (%) | 27.34 | 28.59 | 38.46 | 97.50 | 57.14 | 100 | 100 | 100 | 100 | 100 |
| Hay-windrowed (%) | 100 | 98.35 | 99.57 | 98.69 | 91.02 | 99.56 | 98.47 | 98.69 | 87.12 | 99.34 |
| Oats (%) | 66.21 | 73.78 | 94.85 | 100 | 58.82 | 100 | 100 | 100 | 100 | 100 |
| Soybean-notill (%) | 72.36 | 71.24 | 80.18 | 79.33 | 53.83 | 73.11 | 89.50 | 83.82 | 82.98 | 88.11 |
| Soybean-mintill (%) | 91.85 | 82.31 | 94.38 | 78.94 | 72.81 | 54.74 | 72.98 | 83.61 | 89.40 | 88.38 |
| Soybean-clean (%) | 75.78 | 57.59 | 82.56 | 78.25 | 43.46 | 63.18 | 95.64 | 87.09 | 97.91 | 93.40 |
| Wheat (%) | 99.78 | 99.47 | 100 | 99.14 | 98.90 | 98.92 | 98.92 | 99.46 | 100 | 99.68 |
| Woods (%) | 99.57 | 99.50 | 99.74 | 94.18 | 93.14 | 82.09 | 94.14 | 99.20 | 98.96 | 99.81 |
| Buildings-Grass-Trees-Drives (%) | 83.83 | 92.84 | 97.97 | 85.74 | 73.10 | 65.57 | 90.16 | 89.89 | 99.73 | 99.07 |
| Stone-Steel-Towers (%) | 99.19 | 96.35 | 98.12 | 98.63 | 87.18 | 98.63 | 100 | 98.63 | 98.63 | 99.18 |
| AA (%) | 78.58 | 80.82 | 88.72 | 89.91 | 67.50 | 81.70 | 92.55 | 94.16 | 94.66 | 95.99 |
| OA (%) | 83.56 | 83.01 | 90.78 | 83.91 | 67.20 | 69.95 | 84.36 | 89.94 | 90.88 | 92.47 |
| Kappa | 0.8139 | 0.8071 | 0.8950 | 0.8177 | 0.6325 | 0.6616 | 0.8234 | 0.8855 | 0.8959 | 0.9143 |
| t(s) | 0.98 | 0.34 | 35.80 | 372.96 | 1500.03 | 1251.78 | 3238.74 | 250.16 | 4.81 | 420.02 |
| Surface Object | SVM [6] | ELM [8] | SPELM [9] | SSG [23] | CNN-PPF [16] | BASS-Net [17] | R-VCANet [19] | HiFi-We [29] | BLS [20] | SBLS |
|---|---|---|---|---|---|---|---|---|---|---|
| Brocoli_green_weeds_1 (%) | 100 | 100 | 100 | 98.06 | 99.95 | 99.50 | 99.60 | 99.66 | 99.97 | 100 |
| Brocoli_green_weeds_2 (%) | 99.80 | 100 | 99.95 | 93.84 | 98.84 | 99.65 | 99.87 | 99.19 | 99.51 | 99.81 |
| Fallow (%) | 91.07 | 99.80 | 99.88 | 88.20 | 78.47 | 99.49 | 98.06 | 99.06 | 100 | 99.96 |
| Fallow_rough_plow (%) | 97.33 | 97.01 | 98.87 | 94.29 | 95.81 | 98.84 | 98.91 | 99.07 | 99.33 | 99.80 |
| Fallow_smooth (%) | 97.26 | 91.77 | 96.82 | 90.32 | 96.21 | 97.03 | 99.32 | 98.59 | 98.98 | 99.13 |
| Stubble (%) | 99.70 | 99.97 | 99.98 | 94.54 | 99.61 | 99.80 | 98.65 | 99.20 | 99.78 | 99.80 |
| Celery (%) | 98.26 | 99.90 | 99.81 | 92.89 | 97.66 | 99.72 | 98.20 | 98.73 | 99.53 | 99.84 |
| Grapes_untrained (%) | 85.31 | 77.57 | 86.12 | 56.65 | 72.84 | 65.18 | 70.71 | 78.99 | 88.81 | 91.31 |
| Soil_vinyard_develop (%) | 99.20 | 98.43 | 98.50 | 89.73 | 99.08 | 98.61 | 99.74 | 99.87 | 99.97 | 99.65 |
| Corn_senesced_green_weeds (%) | 84.84 | 95.92 | 96.61 | 77.38 | 80.84 | 87.78 | 91.22 | 89.13 | 93.52 | 94.12 |
| Lettuce_romaine_4wk (%) | 86.68 | 92.56 | 96.49 | 89.43 | 63.20 | 92.18 | 98.66 | 97.73 | 99.54 | 99.79 |
| Lettuce_romaine_5wk (%) | 97.27 | 97.76 | 95.56 | 95.02 | 91.72 | 98.53 | 100 | 99.97 | 99.94 | 100 |
| Lettuce_romaine_6wk (%) | 96.52 | 88.63 | 96.41 | 91.58 | 96.84 | 96.21 | 99.44 | 96.58 | 99.11 | 99.00 |
| Lettuce_romaine_7wk (%) | 86.62 | 76.20 | 88.69 | 89.60 | 88.61 | 96.95 | 97.33 | 96.50 | 97.18 | 97.10 |
| Vinyard_untrained (%) | 67.10 | 81.56 | 79.71 | 72.97 | 60.14 | 67.43 | 74.66 | 87.14 | 82.75 | 89.27 |
| Vinyard_vertical_trellis (%) | 99.24 | 99.93 | 99.98 | 88.35 | 96.74 | 98.15 | 99.44 | 95.86 | 98.68 | 98.80 |
| AA (%) | 92.89 | 93.56 | 95.84 | 87.68 | 88.53 | 93.44 | 95.23 | 95.95 | 97.28 | 97.96 |
| OA (%) | 89.12 | 90.73 | 93.29 | 81.21 | 84.76 | 86.77 | 89.42 | 92.56 | 94.67 | 96.14 |
| Kappa | 0.8793 | 0.8965 | 0.9252 | 0.7927 | 0.8306 | 0.8533 | 0.8824 | 0.9174 | 0.9406 | 0.9570 |
| t(s) | 3.26 | 1.68 | 131.60 | 156.20 | 1560.19 | 1294.50 | 17,080.47 | 352.24 | 13.95 | 240.26 |
| Surface Object | SVM [6] | ELM [8] | SPELM [9] | SSG [23] | CNN-PPF [16] | BASS-Net [17] | R-VCANet [19] | HiFi-We [29] | BLS [20] | SBLS |
|---|---|---|---|---|---|---|---|---|---|---|
| Water (%) | 100 | 99.84 | 100 | 100 | 99.21 | 100 | 100 | 100 | 100 | 98.32 |
| Hippo grass (%) | 88.89 | 93.33 | 97.83 | 87.90 | 100 | 100 | 100 | 96.05 | 99.26 | 97.28 |
| Floodplain grasses1 (%) | 95.43 | 98.5 | 99.57 | 98.35 | 100 | 99.57 | 100 | 96.62 | 100 | 100 |
| Floodplain grasses2 (%) | 91.26 | 78.09 | 95.81 | 96.21 | 94.20 | 97.44 | 100 | 99.49 | 99.28 | 100 |
| Reeds1 (%) | 89.19 | 94.79 | 93.8 | 79.52 | 89.02 | 86.35 | 96.79 | 90.84 | 93.57 | 96.87 |
| Riparian (%) | 62.43 | 100 | 100 | 77.83 | 78.74 | 82.33 | 90.36 | 94.46 | 87.71 | 99.28 |
| Firescar2 (%) | 97.07 | 100 | 100 | 98.74 | 94.35 | 100 | 100 | 95.73 | 100 | 100 |
| Island interior (%) | 97.83 | 98.19 | 98.71 | 97.27 | 87.56 | 100 | 100 | 100 | 100 | 100 |
| Acacia woodlands (%) | 93.40 | 87.69 | 98.47 | 93.47 | 92.09 | 94.22 | 87.07 | 95.78 | 99.25 | 99.86 |
| Acacia shrublands (%) | 75.18 | 88.31 | 99.39 | 90.61 | 93.62 | 96.49 | 99.56 | 98.77 | 100 | 100 |
| Acacia grasslands (%) | 93.85 | 99.02 | 99.93 | 88.14 | 95.40 | 92.63 | 97.54 | 94.95 | 100 | 100 |
| Short mopane (%) | 89.70 | 94.43 | 98.43 | 98.14 | 100 | 100 | 100 | 97.52 | 100 | 100 |
| Mixed mopane (%) | 89.52 | 97.80 | 99.84 | 92.42 | 95.38 | 95.56 | 99.19 | 93.79 | 98.47 | 99.27 |
| Exposed soils (%) | 94.94 | 99.84 | 100 | 97.87 | 100 | 100 | 98.67 | 99.73 | 98.93 | 97.33 |
| AA (%) | 96.51 | 94.61 | 98.70 | 92.61 | 94.26 | 96.04 | 97.80 | 96.70 | 98.32 | 99.16 |
| OA (%) | 96.71 | 94.16 | 98.67 | 92.15 | 93.40 | 95.25 | 97.27 | 96.36 | 98.13 | 99.32 |
| Kappa | 0.9644 | 0.9367 | 0.9856 | 0.9149 | 0.9284 | 0.9485 | 0.9704 | 0.9606 | 0.9798 | 0.9926 |
| t(s) | 1.59 | 1.31 | 12.57 | 16.35 | 1020.09 | 1120.53 | 908.54 | 439.56 | 3.83 | 70.97 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, Y.; Wang, X.; Cheng, Y.; Chen, C.L.P. Hyperspectral Imagery Classification Based on Semi-Supervised Broad Learning System. Remote Sens. 2018, 10, 685. https://doi.org/10.3390/rs10050685
Kong Y, Wang X, Cheng Y, Chen CLP. Hyperspectral Imagery Classification Based on Semi-Supervised Broad Learning System. Remote Sensing. 2018; 10(5):685. https://doi.org/10.3390/rs10050685
Chicago/Turabian StyleKong, Yi, Xuesong Wang, Yuhu Cheng, and C. L. Philip Chen. 2018. "Hyperspectral Imagery Classification Based on Semi-Supervised Broad Learning System" Remote Sensing 10, no. 5: 685. https://doi.org/10.3390/rs10050685





