Tartary Buckwheat Genetic Diversity in the Himalayas Associated with Farmer Landrace Diversity and Low Dietary Dependence
Abstract
:1. Introduction
2. Materials and Methods
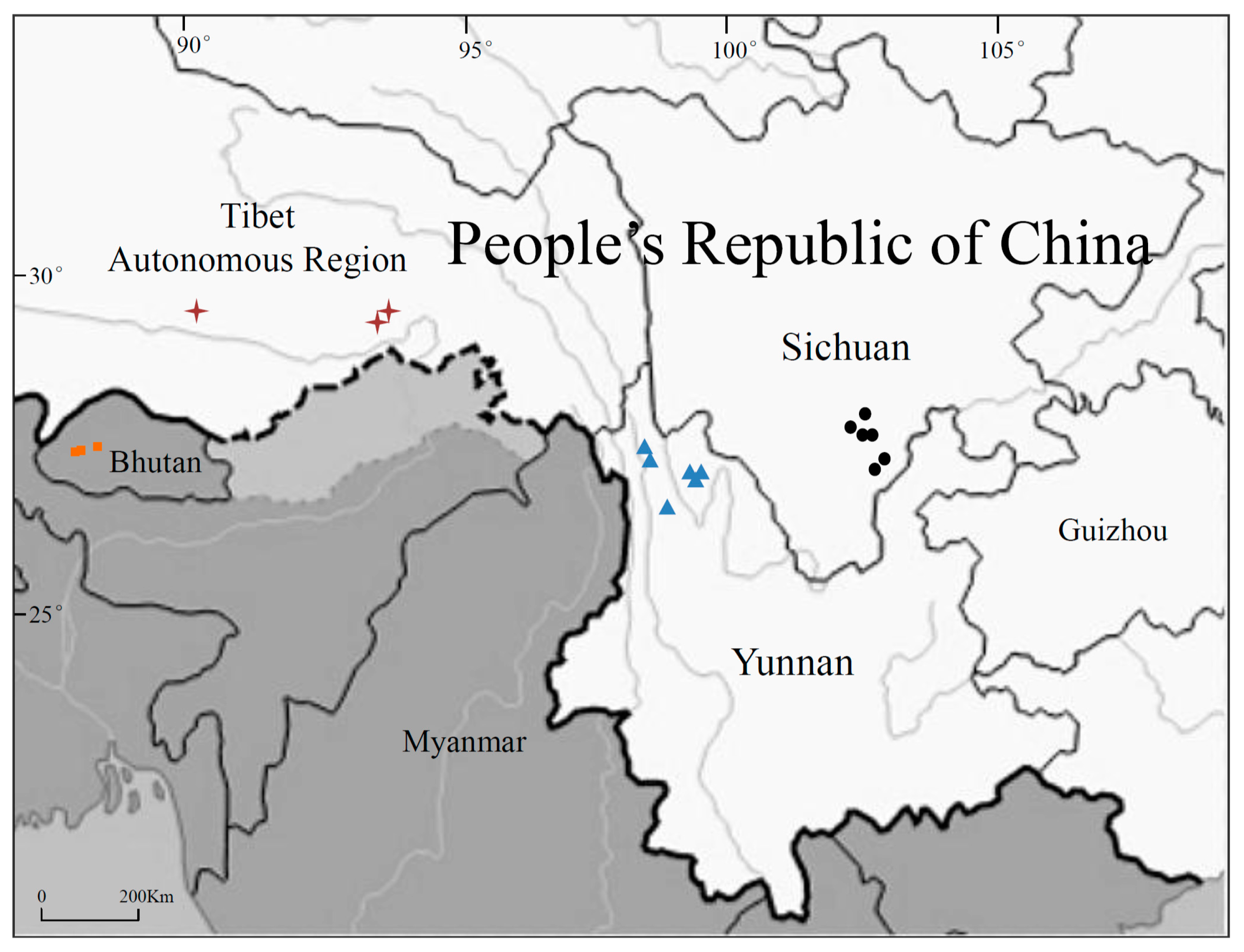
2.1. Study Sites
2.2. Ethnobotanical Surveys
2.3. Sample Collections
2.4. DNA Isolation, PCR Amplification and Sequencing
2.5. Genetic Analysis
Sequence data of F. tataricum samples were analyzed using a series of genetic diversity indexes including gene polymorphisms, nucleotide diversity, and haplotype diversity by MEGA6 and DnaSP5 software [40]. MEGA6 was used for the alignment of multiple sequences and to remove clutter sequences. DnaSP5 was applied to the analysis of nucleotide polymorphisms from aligned DNA sequence data among populations. In order to compare the sequenced locus, a series of genetic diversity indexes including the number of polymorphic sites, nucleotide diversity, average number of nucleotide differences, number of haplotypes, and haplotype (gene) diversity were calculated with MEGA6 and DnaSP 5. Haplotype analyses with and without gaps and indels were performed using DnaSP program. Phylogenetic trees were constructed on the basis of DNA polymorphisms by MEGA6 to distinguish samples from different groups, including the neighbor-joining tree based on single locus trnH-psbA gene as well as the maximum parsimony tree by combining the three study loci for double check. Compared to the other two loci, the trnH-psbA gene provides the highest intraspecies variation and discriminating ability in the chloroplast that has the potential to determine genetic polymorphism.[41]
2.6. Statistical Analysis
3. Results
3.1. Ethnobotanical Survey
3.2. Genetic Analysis
3.3. Correlation Analysis of Cultural, Geographic, and Morphological Factors with Genetic Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hughes, A.R.; Inouye, B.D.; Johnson, M.T.J.; Underwood, N.; Vellend, M. Ecological consequences of genetic diversity. Ecol. Lett. 2008, 11, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Cavatassi, R.; Lipper, L.; Hopkins, J. The Role of Crop Genetic Diversity in Coping with Agricultural Production Shocks: Insights from Eastern Ethiopia; No. 06-17; Food and Agriculture Organization of the United Nations: Rome, Italy, 2006. [Google Scholar]
- Jackson, L.; van Noordwijk, M.; Bengtsson, J.; Foster, W.; Lipper, L.; Pulleman, M.; Said, M.; Snaddon, J.; Vodouhe, R. Biodiversity and agricultural sustainability: From assessment to adaptive manage. Curr. Opin. Environ. Sustain. 2010, 2, 80–87. [Google Scholar] [CrossRef]
- Johns, T.; Powell, B.; Maundu, P.; Eyzaguirre, P.B. Agricultural biodiversity as a link between traditional food systems and contemporary development, social integrity and ecological health. J. Sci. Food Agric. 2013, 93, 3433–3442. [Google Scholar] [CrossRef] [PubMed]
- Rana, R.B.; Garfield, C.; Sthapit, B.; Jarvis, D. Influence of socio-economic and cultural factors in rice varietal diversity management on-farm in Nepal. Agric. Hum. Values 2007, 24, 461–472. [Google Scholar] [CrossRef]
- Jarvis, D.I.; Brown, A.H.; Cuong, P.H.; Collado-Panduro, L.; Latournerie-Moreno, L.; Gyawali, S.; Tanto, T.; Sawadogo, M.; Mar, I.; Sadiki, M.; et al. A global perspective of the richness and evenness of traditional crop-variety diversity maintained by farming communities. Proc. Natl. Acad. Sci. USA 2008, 105, 5326–5331. [Google Scholar] [CrossRef] [PubMed]
- Tuxill, J.; Reyes, L.A.; Latournerie, L.; Cob, V.; Jarvis, D.I. All maize is not Equal: Maize Variety Choices and Mayan Foodways in Rural Yucatan, Mexico. In Pre-Columbian Foodways, Interdisciplinary Approaches to Food, Culture and Markets in Mesoamerica; Staller, J.E., Carrasco, M.D., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 467–486. [Google Scholar]
- Jarvis, D.I.; Hodgkin, T.; Brown, A.H.; Cuong, P.H.; Collado-Panduro, L.; Latournerie-Moreno, L.; Gyawali, S.; Tanto, T.; Sawadogo, M.; Mar, I.; et al. Crop Genetic Diversity in the Field and on the Farm: Principles and Applications in Research Practices; Yale University Press: New Haven, CT, USA, 2016. [Google Scholar]
- Labeyrie, V.; Rono, B.; Leclerc, C. How social organization shapes crop diversity: An ecological anthropology approach among Tharaka farmers of Mount Kenya. AgriC. Hum. Values 2014, 31, 97–107. [Google Scholar] [CrossRef]
- Ahmed, S.; Peters, C.M.; Long, C.L.; Meyer, R.; Unachukwu, U.; Litt, A.; Kennelly, E.; Stepp, J.R. Biodiversity and phytochemical quality in indigenous and state-supported tea management systems of Yunnan, China. Conserv. Lett. 2013, 6, 28–36. [Google Scholar] [CrossRef]
- Altieri, M.A. Linking ecologists and traditional farmers in the search for sustainable agriculture. Front. Ecol. Environ. 2004, 2, 35–42. [Google Scholar] [CrossRef]
- Bisht, I.S.; Mehta, P.S.; Bhandari, D.C. Traditional crop diversity and its conservation on-farm for sustainable agricultural production in Kumaon Himalaya of Uttaranchal state: A case study. Genet. Resour. Crop Evol. 2007, 54, 345–357. [Google Scholar] [CrossRef]
- Jarvis, D.I.; Hodgkin, T.; Sthapit, B.R.; Fadda, C.; Lopez-Noriega, I. A heuristic framework for identifying multiple ways of supporting the conservation and use of traditional crop varieties within the agricultural production system. Crit. Rev. Plant Sci. 2011, 30, 125–176. [Google Scholar] [CrossRef]
- Pardo-de-Santayana, M.; Macía, M.J. Biodiversity: The benefit of traditional knowledge. Nature 2015, 518, 487–488. [Google Scholar] [CrossRef] [PubMed]
- Duc, G.; Bao, S.; Baum, M.; Redden, B.; Sadiki, M.; Suso, M.J.; Vishniakova, M.; Zong, X.X. Diversity maintenance and use of Vicia faba L. genetic resources. Field Crop Res. 2010, 115, 270–278. [Google Scholar] [CrossRef]
- Longley, C. A Social Life of Seeds: Local Management of Crop Variability in North-Western Sierra Leone. Ph.D. Thesis, University of London, London, UK, 2000. [Google Scholar]
- Deu, M.; Sagnard, F.; Chantereau, J.; Calatayud, C.; Hérault, D.; Mariac, C.; Pham, J.-L.; Vigouroux, Y.; Kapran, I.; Traore, P.S.; et al. Nigerwide assessment of in situ sorghum genetic diversity with microsatellite markers. Theor. Appl. Genet. 2008, 116, 903–913. [Google Scholar] [CrossRef] [PubMed]
- Delêtrea, M.; McKey, D.B.; Hodkinson, T.R. Marriage exchanges, seed exchanges, and the dynamics of manioc diversity. Proc. Natl Acad. Sci. USA 2011, 108, 18249–18254. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, C.; d’Eeckenbrugge, G.C. Social organization of crop genetic diversity. The G × E × S interaction model. Diversity 2012, 4, 1–32. [Google Scholar] [CrossRef] [Green Version]
- Ohnishi, O. Discovery of new Fagopyrum species and its implication for the studies of evolution of Fagopyrum and of the origin of cultivated buckwheat. In Current Advances in Buckwheat Research; Matano, T., Ujihara, A., Eds.; Shinshu University Press: Nagano, Japan, 2016; Volume I–III, pp. 175–190. [Google Scholar]
- Jones, M.K.; Liu, X.Y. Origins of agriculture in East Asia. Science 2009, 324, 730. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, K.; Ohnishi, O. Phylogenetic relationships among wild and cultivated tartary buckwheat (Fagopyrum tataricum Gaertn.) populations revealed by AFLP analyses. Genes Genet. Syst. 2001a, 76, 47–52. [Google Scholar] [CrossRef]
- Tsuji, K.; Ohnishi, O. Phylogenetic position of east Tibetan natural populations in tatary buckwheat (Fagopyrum tataricum Gaertn.) revealed by RAPD analyses. Genet. Resour. Crop Evol. 2001, 48, 63–67. [Google Scholar] [CrossRef]
- Senthilkumaran, R.; Bisht, I.S.; Bhat, K.V.; Rana, J.C. Diversity in buckwheat north-western Indian Himalayas. Genet. Resour. Crop Evol. 2008, 55, 287–302. [Google Scholar] [CrossRef]
- Zhou, X.L.; Wen, L.; Li, Z.; Zhou, Y.; Chen, Y.; Lu, Y. Advance on the benefits of bioactive peptides from buckwheat. Phytochem. Rev. 2015, 14, 381–388. [Google Scholar] [CrossRef]
- Wu, D. Buckwheat, globalization and the reproduction of Yi culture. China Agric. Univ. J. (Soc. Sci. Ed.) 2011, 3, 193–197. (In Chinese) [Google Scholar]
- Jarvis, D.I. Pollen evidence of changing Holocene monsoon climate in Sichuan Province, China. Quat. Res. 1993, 39, 325–337. [Google Scholar] [CrossRef]
- Stoeckle, M. Taxonomy, DNA and the barcode of life. BioScience 2003, 53, 2–3. [Google Scholar] [CrossRef]
- Hajibabaei, M.; Singer, G.A.; Hebert, P.D.; Hickey, D.A. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. TRENDS Genet. 2007, 23, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Sun, Z.; Linghu, B.; Xu, D.; Wu, B.; Zhang, B.; Wang, X.; Han, Y.; Zhang, L.; Qiao, Z.; et al. Genetic diversity of buckwheat cultivars (Fagopyrum tartaricum Gaertn.) assessed with SSR markers developed from genome survey sequences. Plant Mol. Biol. Rep. 2016, 34, 233–241. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, L. Molecular characterization of genetic diversity of underutilized crops: Buckwheat as an example. Acta Hort. 2013, 979, 407–419. [Google Scholar] [CrossRef]
- Tsuji, K.; Ohnishi, O. Origin of cultivated Tatary buckwheat (Fagopyrum tataricum Gaertn.) revealed by RAPD analyses. Genet Resour Crop Ev. 2000, 47, 431–438. [Google Scholar]
- Li, C.H.; Kobayashi, K.; Yoshida, Y.; Ohsawa, R. Genetic analyses of agronomic traits in Tartary buckwheat (Fagopyrum tataricum (L.) Gaertn.). Breeding Sci. 2012, 62, 303–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neuhoff, D.; Tashi, S.; Rahmann, G.; Denich, M. Organic agriculture in Bhutan: Potential and challenges. Org. Agric. 2014, 4, 209–221. [Google Scholar]
- Silva, H.C.; Caraciolo, R.L.F.; Marangon, L.C.; Ramos, M.A.; Santos, L.L.; Albuquerque, U.P. Evaluating different methods used in ethnobotanical and ecological studies to record plant biodiversity. J. Ethnobiol. Ethnomedi. 2014, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Kischner, R. Cercosporella and Ramularia. Mycologia 2009, 101, 110. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Hajiahmadi, Z.; Talebi, M.; Sayed-Tabatabaei, B.E. Studying genetic variability of Pomegranate (Punica granatum L.) based on chloroplast DNA. Mol. Biotechnol. 2013, 55, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Brush, S.B. Genes in the Field: On-Farm Conservation of Crop Diversity; International Plant Genetic Resources Institute: Rome, Italy, 2000. [Google Scholar]
- Vigouroux, Y.; Barnaud, A.; Scarcelli, N.; Thuillet, A.C. Biodiversity, evolution and adaptation in cultivated crops. C.R. Biol. 2011, 334, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Coomes, O.T.; McGuire, S.J.; Garine, E.; Caillon, S.; Mckey, D.; Demeulenaere, E.; Javis, D.; Aistara, G.; Barnaud, A.; Clouvel, P.; et al. Farmer seed networks make a limited contribution to agriculture? Four common misconceptions. Food Policy 2015, 56, 41–50. [Google Scholar] [CrossRef] [Green Version]
- Pfeifer, M.; Jetschke, G. Influence of geographical isolation on genetic diversity of Himantoglossum hircinum (Orchidaceae). Folia Geobot. 2006, 41, 3–20. [Google Scholar] [CrossRef]
- Banerjee, A.; Bandopadhyay, R. Biodiversity hotspot of Bhutan and its sustainability. Curr. Sci. 2016, 110, 521–527. [Google Scholar] [CrossRef]
- Idrissi, A.; Ouazzani, N. Contribution of morphological descriptors to the inventory and identification of olive (Olea europaea L.) varieties. Plant Genet. Resour. Newsl. 2003, 136, 1–10. [Google Scholar]
- IAASTD. Agriculture at a Crossroads; McIntyre, B.D., Herren, H.R., Wakhungu, J., Watson, R.T., Eds.; International Assessment of Agricultural Knowledge, Science and Technology for Development (Island Press): Quezon, Philippines, 2009; pp. 65–86. [Google Scholar]
- Chrungoo, N.K.; Sangma, S.C.; Bhatt, V.; Raina, S.N. Wild Crop Relatives: Genomic and Breeding Resources; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 293–307. [Google Scholar]
- Hodgkin, T.; Rana, R.; Tuxill, J.; Balma, D.; Subedi, A.; Mar, I.; Karamura, D.; Valdivia, R.; Collado, L.; Latournerie, L.; et al. Seed Systems and Crop Genetic Diversity in Agroecosystems; Jarvis, D.I., Padoch, C., Cooper, D., Eds.; Managing Biodiversity in Agricultural Ecosystems (Columbia University Press): New York, NY, USA, 2007; pp. 77–116. [Google Scholar]
- Sperling, L. When Disaster Strikes: A Guide for Assessing Seed Security; CIAT: Cali, Colombia, 2008. [Google Scholar]
- Li, Y.H.; Dong, C.Y. On amendment and improvement of the PRC seed law: From the perspective of variety certification system. Law Sci. Mag. 2014, 12, 57–64. (In Chinese) [Google Scholar]
- Pan, Q.Y.; Huang, B.L.; Vernooy, R.; Song, Y.Q. Participatory plant breeding promotes variety improvement and biodiversity. Guizhou Agric. Sci. 2004, 32, 80–81. (In Chinese) [Google Scholar]
 ,
,  ,
,  and
and  represent samples from Sichuan, Yunnan, Tibet and Bhutan respectively. A sample of Fagopyrum esculentum collected from Inner Mongolia as well as F. dibotrys and F. tibeticum downloaded from Genbank served as outgroups). This phylogenetic tree based on the trnH-psbA gene indicates that buckwheat samples from Sichuan had fewer clades and lower genetic diversity compared with samples from the other sites.
represent samples from Sichuan, Yunnan, Tibet and Bhutan respectively. A sample of Fagopyrum esculentum collected from Inner Mongolia as well as F. dibotrys and F. tibeticum downloaded from Genbank served as outgroups). This phylogenetic tree based on the trnH-psbA gene indicates that buckwheat samples from Sichuan had fewer clades and lower genetic diversity compared with samples from the other sites.
 ,
,  ,
,  and
and  represent samples from Sichuan, Yunnan, Tibet and Bhutan respectively. A sample of Fagopyrum esculentum collected from Inner Mongolia as well as F. dibotrys and F. tibeticum downloaded from Genbank served as outgroups). This phylogenetic tree based on the trnH-psbA gene indicates that buckwheat samples from Sichuan had fewer clades and lower genetic diversity compared with samples from the other sites.
represent samples from Sichuan, Yunnan, Tibet and Bhutan respectively. A sample of Fagopyrum esculentum collected from Inner Mongolia as well as F. dibotrys and F. tibeticum downloaded from Genbank served as outgroups). This phylogenetic tree based on the trnH-psbA gene indicates that buckwheat samples from Sichuan had fewer clades and lower genetic diversity compared with samples from the other sites.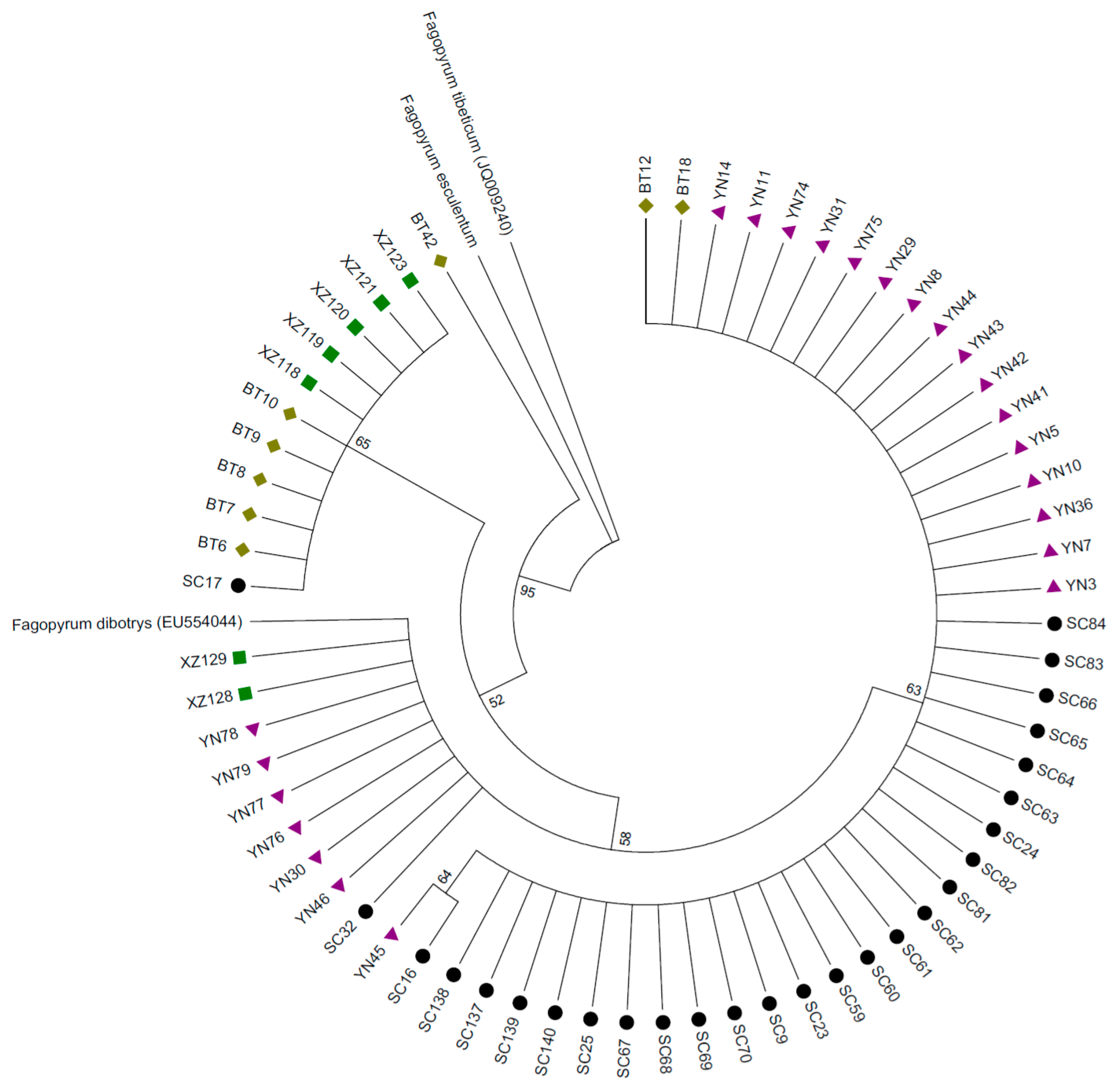
 ,
,  ,
,  and
and  represent samples from Sichuan, Yunnan, Tibet and Bhutan respectively. A sample of Fagopyrum esculentum collected from Inner Mongolia as well as Fagopyrum dibotrys and Fagopyrum tibeticum downloaded from Genbank served as outgroups). This phylogenetic tree based on combining trnH-psbA, rbcL-a and trnL-F genes indicates that buckwheat samples from Sichuan had fewer clades and lower genetic diversity compared with samples from the other sites.
represent samples from Sichuan, Yunnan, Tibet and Bhutan respectively. A sample of Fagopyrum esculentum collected from Inner Mongolia as well as Fagopyrum dibotrys and Fagopyrum tibeticum downloaded from Genbank served as outgroups). This phylogenetic tree based on combining trnH-psbA, rbcL-a and trnL-F genes indicates that buckwheat samples from Sichuan had fewer clades and lower genetic diversity compared with samples from the other sites.
 ,
,  ,
,  and
and  represent samples from Sichuan, Yunnan, Tibet and Bhutan respectively. A sample of Fagopyrum esculentum collected from Inner Mongolia as well as Fagopyrum dibotrys and Fagopyrum tibeticum downloaded from Genbank served as outgroups). This phylogenetic tree based on combining trnH-psbA, rbcL-a and trnL-F genes indicates that buckwheat samples from Sichuan had fewer clades and lower genetic diversity compared with samples from the other sites.
represent samples from Sichuan, Yunnan, Tibet and Bhutan respectively. A sample of Fagopyrum esculentum collected from Inner Mongolia as well as Fagopyrum dibotrys and Fagopyrum tibeticum downloaded from Genbank served as outgroups). This phylogenetic tree based on combining trnH-psbA, rbcL-a and trnL-F genes indicates that buckwheat samples from Sichuan had fewer clades and lower genetic diversity compared with samples from the other sites.

| Location | Study Site | Population Code | Number of Plants | Geographic Coordinates | Altitude (m) | Climate | Topography | Main Sociolinguistic Group | Major Staple Food | Cultural Lifestyle | Religion |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sichuan | Zhaojue | 1 | 6 | 28°00′51.18′′ N 102°50′38.11′′E | 3010 | subtropical monsoon | mountain plateau | Yi | tartary buckwheat | farming | Bimoism |
| Zhaojue | 2 | 6 | 28°03′18.17′′ N 102°49′54.83′′E | 2570 | subtropical monsoon | mountain plateau | Yi | tartary buckwheat | farming | Bimoism | |
| Zhaojue | 3 | 6 | 27°50′26.11′′ N 102°47′08.91′′E | 2260 | subtropical monsoon | mountain plateau | Yi | tartary buckwheat | farming | Bimoism | |
| Zhaojue | 4 | 6 | 27°54′03.10′′ N 102°50′18.16′′E | 2980 | subtropical monsoon | mountain plateau | Yi | tartary buckwheat | farming | Bimoism | |
| Butuo | 5 | 5 | 27°42′41.91′′ N 102°49′06.09′′E | 2450 | subtropical monsoon | mountain plateau | Yi | tartary buckwheat | farming | Bimoism | |
| Butuo | 6 | 5 | 27°44′26.78′′ N 102°47′01.00′′E | 2460 | subtropical monsoon | mountain plateau | Yi | tartary buckwheat | farming | Bimoism | |
| Yunnan | Weixi | 7 | 5 | 27°10′39.00′′ N 99°17′57.75′′E | 2590 | cold temperate | mountain ravines | Tibetan | hulless barley | nomadic | Buddhism |
| Deqin | 8 | 5 | 28°29′02.26′′ N 98°56′32.30′′E | 4360 | cold temperate | mountain ravines | Tibetan | hulless barley | nomadic | Buddhism | |
| Deqin | 9 | 5 | 28°27′06.58′′ N 98°54′23.39′′E | 3280 | cold temperate | mountain ravines | Tibetan | hulless barley | nomadic | Buddhism | |
| Shangri-La | 10 | 6 | 27°49′37.07′′ N 99°42′25.15′′E | 3280 | cold temperate | mountain ravines | Tibetan | hulless barley | nomadic | Buddhism | |
| Shangri-La | 11 | 6 | 27°43′19.63′′ N 99°46′06.74′′E | 3920 | cold temperate | mountain ravines | Tibetan | hulless barley | nomadic | Buddhism | |
| Shangri-La | 12 | 6 | 27°28′51.87′′ N 99°52′52.37′′E | 3500 | cold temperate | mountain ravines | Tibetan | hulless barley | nomadic | Buddhism | |
| Tibet | Lhasa | 13 | 5 | 29°38′46.80′′ N 91°08′25.02′′E | 3650 | plateau temperate | Tibetan plateau | Tibetan | hulless barley | nomadic | Buddhism |
| Linzhi | 14 | 5 | 29°38′57.02′′ N 94°21′41.70′′E | 3000 | plateau temperate | Tibetan plateau | Tibetan | hulless barley | nomadic | Buddhism | |
| Milin | 15 | 5 | 29°12′57.47′′ N 94°12′49.08′′E | 2940 | plateau temperate | Tibetan plateau | Tibetan | hulless barley | nomadic | Buddhism | |
| Bhutan | Thimphu | 16 | 10 | 27°28′21.79′′ N 89°38′20.70′′E | 2340 | wet monsoonal | mountain valley | Bhutanese | wheat | farming | Buddhism |
| Paro | 17 | 10 | 27°25′45.30 N 89°24′59.72′′E | 2270 | wet monsoonal | mountain valley | Bhutanese | wheat | farming | Buddhism | |
| Paro | 18 | 10 | 27°27′08.94 N 89°26′01.41′′E | 2410 | wet monsoonal | mountain valley | Bhutanese | wheat | farming | Buddhism |
| *Loci (Avg. Sequence Length) | **Groups | Number of Amplified Sequences | Number of Polymorphic Sites | Nucleotide Diversity (Pi) | Avg. Number of Nucleotide Differences (K) | Number of Haplotypes (NHap) | Haplotype (Gene) Diversity (Hd) | Genbank No. |
|---|---|---|---|---|---|---|---|---|
| rbcL-a (633 bp) | SC | 29 | 4 | 0.00138 | 0.857 | 4 | 0.362 | Serial No.: KP966643, KP966644, KP966645, KP966646 … KP966728 |
| YN | 28 | 8 | 0.00295 | 1.968 | 6 | 0.712 | ||
| XZ | 5 | 3 | 0.00206 | 1.400 | 4 | 0.900 | ||
| BT | 26 | 5 | 0.00199 | 1.262 | 4 | 0.668 | ||
| trnH-psbA (408 bp) | SC | 27 | 3 | 0.00080 | 0.291 | 4 | 0.214 | Serial No.: KP966756, KP966757, KP966758, KP966759 … KP966820 |
| YN | 23 | 5 | 0.00251 | 0.901 | 5 | 0.561 | ||
| XZ | 7 | 2 | 0.00180 | 0.762 | 3 | 0.524 | ||
| BT | 8 | 2 | 0.00249 | 0.964 | 3 | 0.607 | ||
| trnL-F (849 bp) | SC | 5 | 1 | 0.00047 | 0.400 | 2 | 0.400 | Serial No.: KP966729, KP966730, KP966731, KP966732 … KP966755 |
| YN | 6 | 3 | 0.00186 | 1.533 | 3 | 0.733 | ||
| XZ | 3 | 2 | 0.00152 | 1.333 | 2 | 0.667 | ||
| BT | 13 | 2 | 0.00116 | 1.026 | 2 | 0.513 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, W.; Jarvis, D.I.; Ahmed, S.; Long, C. Tartary Buckwheat Genetic Diversity in the Himalayas Associated with Farmer Landrace Diversity and Low Dietary Dependence. Sustainability 2017, 9, 1806. https://doi.org/10.3390/su9101806
Huang W, Jarvis DI, Ahmed S, Long C. Tartary Buckwheat Genetic Diversity in the Himalayas Associated with Farmer Landrace Diversity and Low Dietary Dependence. Sustainability. 2017; 9(10):1806. https://doi.org/10.3390/su9101806
Chicago/Turabian StyleHuang, Weijuan, Devra I. Jarvis, Selena Ahmed, and Chunlin Long. 2017. "Tartary Buckwheat Genetic Diversity in the Himalayas Associated with Farmer Landrace Diversity and Low Dietary Dependence" Sustainability 9, no. 10: 1806. https://doi.org/10.3390/su9101806
APA StyleHuang, W., Jarvis, D. I., Ahmed, S., & Long, C. (2017). Tartary Buckwheat Genetic Diversity in the Himalayas Associated with Farmer Landrace Diversity and Low Dietary Dependence. Sustainability, 9(10), 1806. https://doi.org/10.3390/su9101806






