Analysis of Soil Properties, Bacterial Community Composition, and Metabolic Diversity in Fluvisols of a Floodplain Area
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Information
2.2. Sample Collection
2.3. Analysis of Soil Physico–Chemical Properties
2.4. Analysis of Soil Biological Properties
2.5. Physiological Profiling of Microbial Communities—Biolog EcoPlate™
2.6. Soil DNA Extraction and Next Generation Sequencing (Miseq, Illumina)
2.7. Statistical Analysis
3. Results
3.1. Soil Physico–Chemical Properties
3.2. Soil Biological Properties
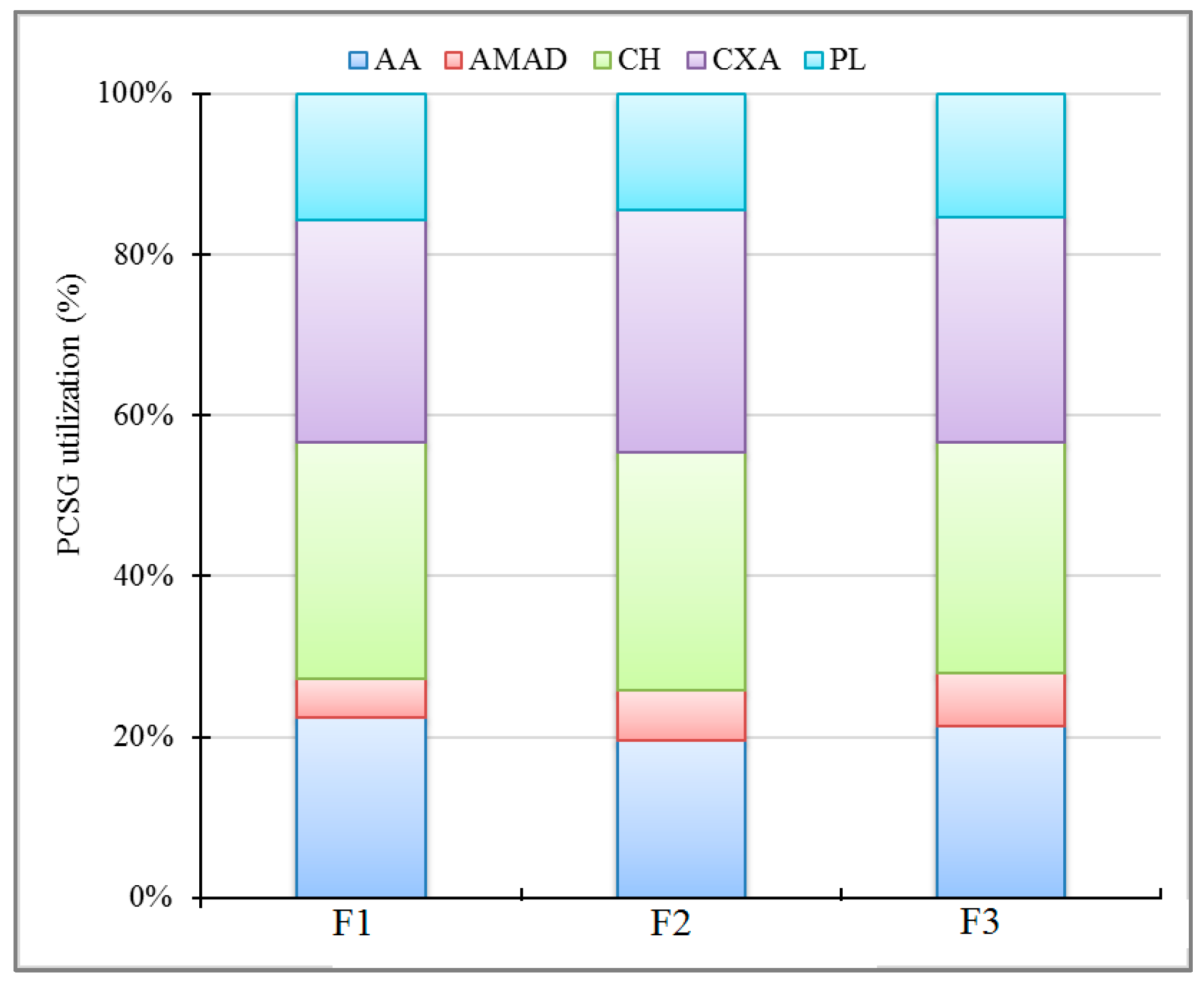
3.3. Microbial Metabolic Potential
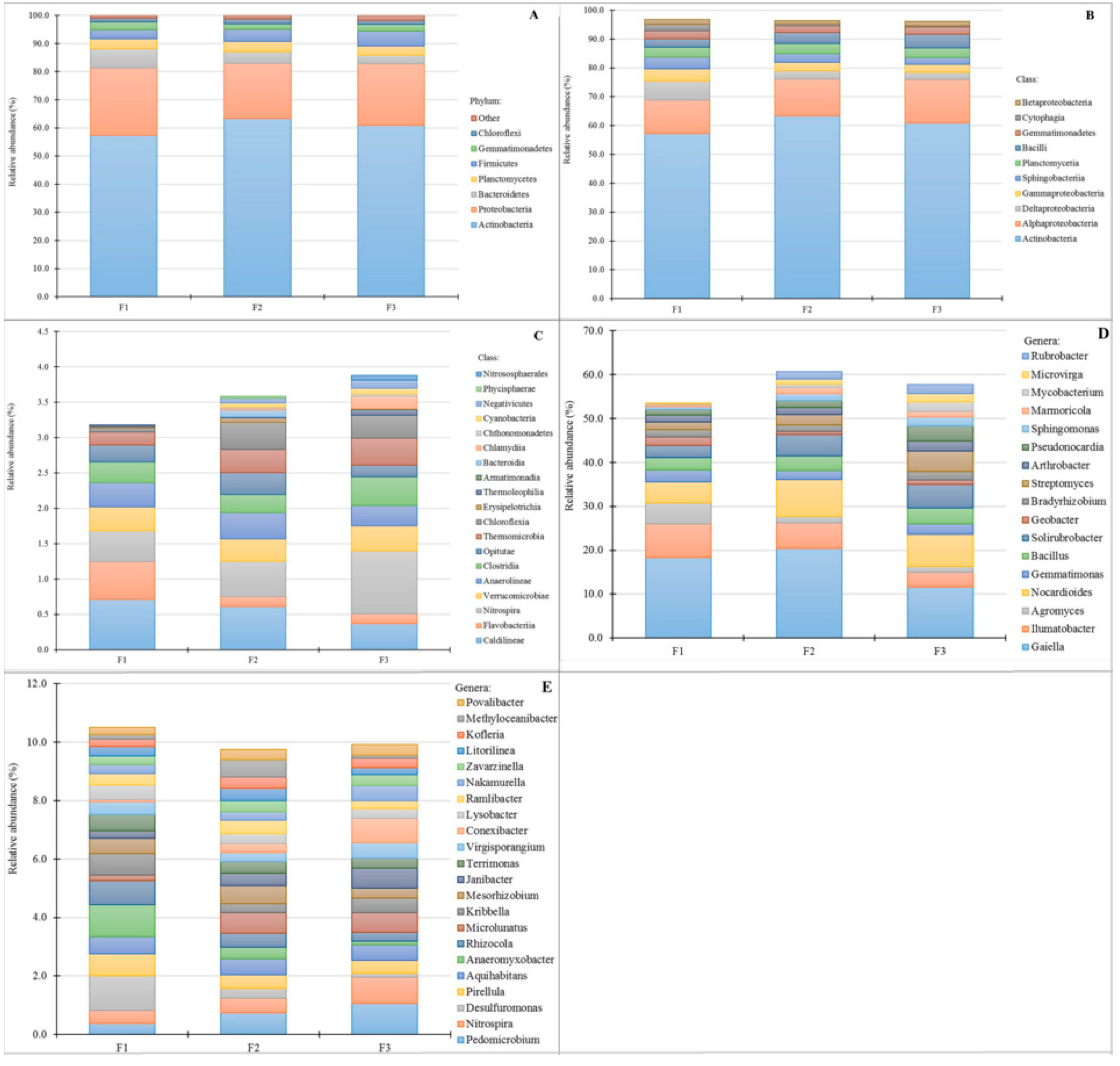
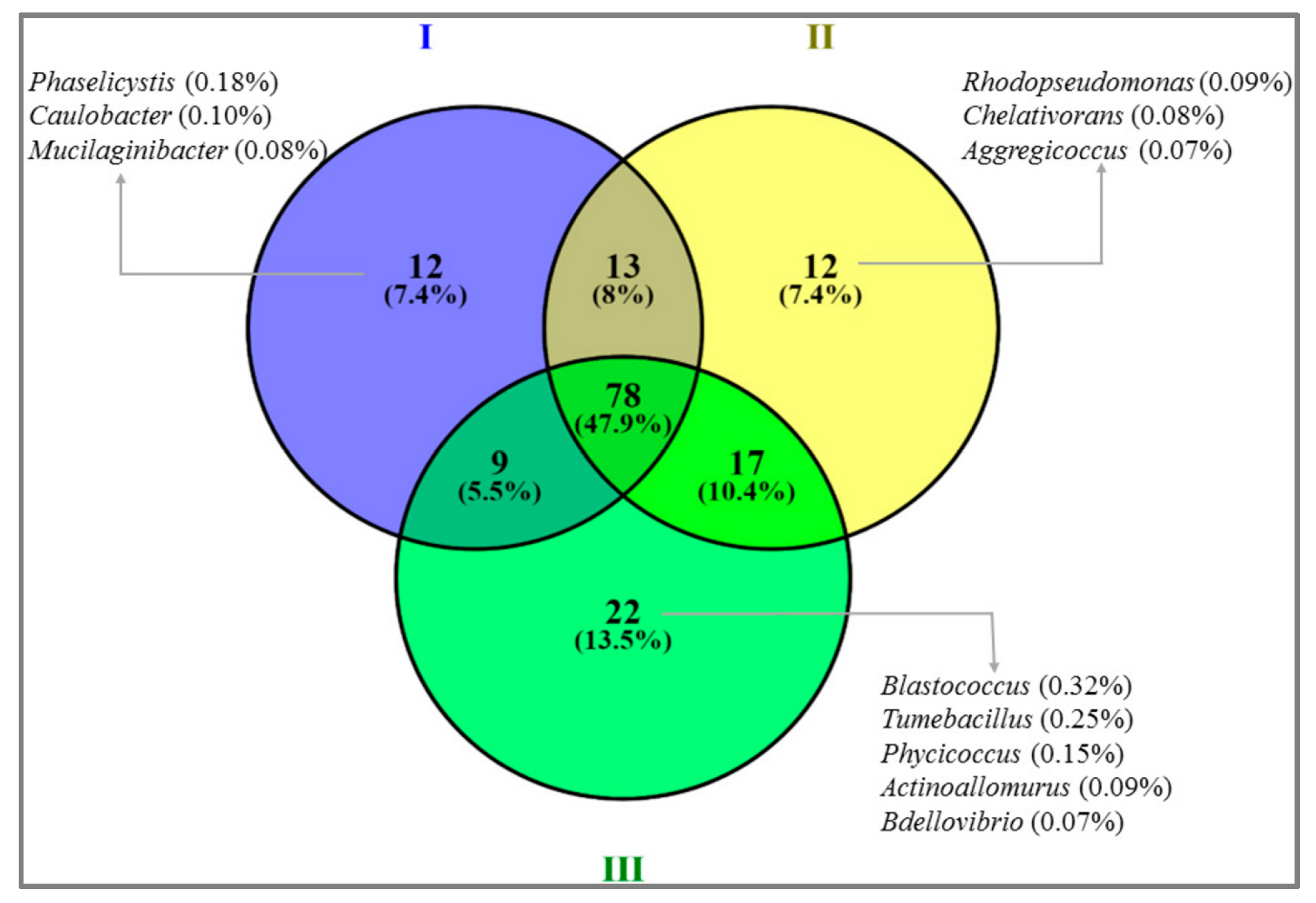
3.4. Bacterial Community Abundance and Composition
3.5. Correlation of Soil Properties and the Bacterial Community
4. Discussion
4.1. Soil Physicochemical Properties
4.2. Soil Biological Properties
4.3. Microbial Metabolic Potential and Bacterial Community Composition
4.4. Relationships between Bacterial Community and Physicochemical and Biological Parameters of Fluvisols
4.5. What Can Be Responsible for Differences between Microbiomes in the Examined Fluvisoils?
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Li, P.; Zhang, T.; Wang, X.; Yu, D. Development of biological soil quality indicator system for subtropical China. Soil Tillage Res. 2013, 126, 112–118. [Google Scholar] [CrossRef]
- Lemaire, G.; Franzluebbers, A.; Carvalho, P.C.; Dedieu, B. Integrated crop-livestock system: Strategies to achieve synergy between agricultural production and environmental quality. Agric. Ecosyst. Environ. 2014, 190, 4–8. [Google Scholar] [CrossRef]
- Handelsman, J.; Rondon, M.R.; Brady, S.F.; Clardy, J.; Goodman, R.M. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem. Biol. 1998, 5, 245–249. [Google Scholar] [CrossRef]
- Kozdrój, J. Metagenome—A new source of information about soil microorganisms. Postępy Mikrobiologii 2013, 52, 185–200. (In Polish) [Google Scholar]
- Nannipieri, P.; Ascher, J.; Ceccherini, M.T.; Landi, L.; Pietramellara, G.; Renella, G. Microbial diversity and soil functions. Eur. J. Soil Sci. 2003, 54, 655–670. [Google Scholar] [CrossRef]
- Grządziel, J.; Gałązka, A. Microplot long-term experiment reveals strong soil type influence on bacteria composition and its functional diversity. Appl. Soil Ecol. 2018, 124, 117–123. [Google Scholar] [CrossRef]
- Gałązka, A.; Furtak, K. Functional microbial diversity in context to agriculture. In Advances in Microbial Diversity in Genomic Era; Das, S., Dash, H.R., Eds.; Academic Press, Elsevier Inc.: Cambridge, MA, USA, 2019; pp. 347–358. [Google Scholar] [CrossRef]
- Furtak, K.; Gajda, A.M. Biochemical methods for the evaluation of the functional and structural diversity of microorganisms in the soil environment. Postępy Mikrobiologii 2018, 57, 194–202. (In Polish) [Google Scholar]
- Gajda, A.M.; Furtak, K. Measuring the Effects of Farming Systems on Physical, Chemical and Microbiological Parameters of Soil Quality. In Novel Methods and Results of Landscape Research in Europe, Central Asia and Siberia Vol. 1, Landscapes in the 21th Century: Status Analyses. Basic Processes and Research Concepts; Sychev, V.G., Mueller, L., Eds.; Publishing House FSBSI “Pryanishnikov Institute of Agrochemistry”: Moscow, Russia, 2018; pp. 212–217. [Google Scholar] [CrossRef]
- Gałązka, A.; Łyszcz, M.; Abramczyk, B.; Furtak, K.; Grządziel, J.; Czaban, J.; Pikulicka, A. Biodiversity of soil environment—Overview of parameters and methods in soil biodiversity analyses. In Proceedings of the Monografie i Rozprawy Naukowe IUNG-PIB, Puławy, Poland, 14–15 September 2016. (In Polish). [Google Scholar]
- Harasimuk, M.; Domonik, A.; Machalski, M.; Pinińska, J.; Warowna, J.; Szymkowiak, A. Małopolska Gap of Vistula River—Projected geopark. Prz. Geol. 2011, 59, 405–416. (In Polish) [Google Scholar]
- Kot, J.; Kucharczyk, M. Pilot Programmes for Management of Natura 2000 Areas “Lesser Poland Gorge of the Vistula”, “Małopolska Gap of Vistula” and “Valley of Zwolenka”; Infrastruktura i ekologia terenów wiejskich, Polska Akademia Nauk, Oddział w Krakowie: Krakow, Poland, 2006; pp. 55–66. (In Polish) [Google Scholar]
- Natura 2000. Available online: http://natura2000.eea.europa.eu/ (accessed on 11 January 2019).
- Malinowski, R. Charecteristic of physical and oxy-redox properties of some alluvial soils of the Cedynia Landscape Park Polder. Folia Univ. Agric. Stetin. 2007, 259, 91–102. (In Polish) [Google Scholar]
- Banach-Szott, M.; Kondratowicz-Maciejewska, K.; Kobierski, M. Humic substances in Fluvisols of the Lower Vistula floodplain, North Poland. Environ. Sci. Pollut. Res. 2018, 25, 23992–24002. [Google Scholar] [CrossRef]
- Özcan, H. Fluvisols. In The Soils of Turkey; World Soils Book Series; Kapur, S., Akça, E., Günal, H., Eds.; Springer International Publishing AG: Cham, Switzerland, 2018; pp. 129–137. [Google Scholar]
- Climate-Data.org. Available online: https://pl.climate-data.org (accessed on 11 January 2019).
- Litynski, T.; Jurkowska, H.; Gorlach, E. Chemical and Agricultural Analysis; Wydawnictwo Naukowe PWN: Warszawa, Polska, 1976. (In Polish) [Google Scholar]
- Czyż, E.A.; Dexter, A.R.; Terelak, H. Content of readily-dispersible clay in the arable layer of some Polish soils. Adv. Geoecol. 2002, 35, 115–124. [Google Scholar]
- DIN/ISO 13878:1998. Soil Quality—Determination of Total Nitrogen Content by Dry Combustion (“Elemental Analysis”); American National Standards Institute: Washington, DC, USA, 2007. [Google Scholar]
- Ostrowska, A.; Gawliński, S.; Szczubiałka, Z. Methods of Analysis and Assessment of Soil and Plant Properties. Catalogue; Dział wydawnictw IOŚ: Warszawa, Poland, 1991. (In Polish) [Google Scholar]
- Casida, L.; Klein, D.; Santoro, T. Soil Dehydrogenase Activity. Soil Sci. 1964, 98, 371–376. [Google Scholar] [CrossRef]
- Tabatabai, M.A. Soil enzymes. In Methods of Soil Analysis; Part 2; Page, A.L., Miller, R.H., Keeney, D.R., Eds.; American Society of Agronomy and Soil Science Society of America: Madison, WI, USA, 1982. [Google Scholar]
- PN-ISO 1038-6:1998. Soil Quality—Determination of Soil Microbial Biomass—Part 2: Fumigation-Extraction Method; American National Standards Institute: Washington, DC, USA, 2007. [Google Scholar]
- Weber, K.P.; Legge, R.L. One-dimensional metric for tracking bacterial community divergence using sole carbon source utilization patterns. J. Microbiol. Methods 2009, 79, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Insam, H.; Goberna, M. Use of Biolog R for community level physiological profiling (CLPP) of environmental samples. In Molecular Microbial Ecology Manual, 2nd ed.; Kowalchuk, G.A., de Brujin, F.J., Head, I.M., Akkermans, A.D., van Elsas, J.D., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2004; pp. 853–860. [Google Scholar]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing. 2016. Available online: https://www.r-project.org/ (accessed on 5 February 2019).
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Garland, J.L.; Millis, A.L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl. Environ. Microbiol. 1991, 57, 2351–2359. [Google Scholar]
- Hill, T.C.J.; Walsh, K.A.; Harris, J.A.; Moffett, B.F. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 2003, 43, 1–11. [Google Scholar] [CrossRef]
- Zak, J.; Willig, M.; Moorhead, D.; Wildman, H. Functional diversity of microbial communities: A quantitative approach. Soil Biol. Biochem. 1994, 26, 1101–1108. [Google Scholar] [CrossRef]
- Jałowiecki, L.; Chojniak, J.M.; Dorgeloh, E.; Hegedusova, B.; Ejhed, H.; Magner, J.; Plaza, G.A. Microbial Community Profiles in Wastewaters from Onsite Wastewater Treatment Systems Technology. PLoS ONE 2016, 11, e0147725. [Google Scholar] [CrossRef]
- Particle size distribution and textural classes of soils and mineral materials—Classification of Polish Society of Soil Science 2008. Roczniki Gleboznawcze Soil Sci. Annu. 2008, 60, 5–16.
- Nannipieri, P.; Grego, S.; Ceccanti, B. Ecological Significance of the Biological Activity in Soils; Marcel Dekker: New York, NY, USA, 1990. [Google Scholar]
- Preston-Mafham, J.; Boddy, L.; Randerson, P.F. Analysis of microbial community functional diversity using sole-carbon-source utilisation profiles—A critique. FEMS Microbiol. Ecol. 2002, 42, 1–14. [Google Scholar] [CrossRef]
- Derakshani, M.; Lukow, T.; Liesack, W. Novel bacterial lineages at the (sub)division level as detected by signature nucleotide-targeted recovery of 16S rRNA genes from bulk soil and rice roots of flooded rice microcosms. Appl. Environ. Microbiol. 2001, 67, 623–631. [Google Scholar] [CrossRef]
- Kobierski, M. Evaluation of the content of heavy metals in fluvisols of floodplain area depending on the type of land use. J. Ecol. Eng. 2015, 16, 23–31. [Google Scholar] [CrossRef]
- Journal of Laws 2002 No 165, item 1359; Dziennik Ustaw 2002. Rozporządzenie Ministra Środowiska, Żelichowski, S. z dn. 09.09.2002 w sprawie standardów jakości gleby oraz jakości ziemi. Dz. U. 2002, Nr 165, poz. 1359. Available online: http://prawo.sejm.gov.pl/isap.nsf/DocDetails.xsp? id=WDU200216 51359 (accessed on 20 May 2019).
- Beena, K.N.; Jaya, D.S. Evaluation of soil contamination in the surroundings of Kerala Minerals and Metals Limited (KMML) industrial area in Kollam District, Kerala, South India. J. Soil Sci. Environ. Manag. 2016, 7, 92–99. [Google Scholar] [CrossRef] [Green Version]
- Sowiński, P.; Glińska-Lewczuk, K.; Kalisz, B.; Astel, A. Distribution of heavy metals in soils in a postglacial river valley—A geochemical landscape approach. Environ. Eng. Manag. J. 2016, 15, 1323–1335. [Google Scholar] [CrossRef]
- Ou, Y.; Rousseau, A.N.; Wang, L.; Yan, B.; Gumiere, T.; Zhu, H. Identification of the alteration of riparian wetland on soil properties, enzyme activities and microbial communities following extreme flooding. Geoderma 2019, 337, 825–833. [Google Scholar] [CrossRef]
- Liu, B.; Huang, Q.; Su, Y.; Wang, M.; Kelly, R.M.; Sun, L. Speciation of nickel and enzyme activities in fluvo-aquic soil under organic amendments treatment. Soil Res. 2018, 56, 456–467. [Google Scholar] [CrossRef]
- Shuler, M.; Kargi, F. Bioprocess Engineering Basic Concepts; Prentice-Hall Incorporation: Englewood Cliffs, NJ, USA, 2010. [Google Scholar]
- Goenster, S.; Gründler, C.; Buerkert, A.; Joergensen, R.G. Soil microbial indicators across land use types in the river oasis Bulgan sum center, Western Mongolia. Ecol. Indic. 2017, 76, 111–118. [Google Scholar] [CrossRef]
- Acosta-Martínez, V.; Acosta-Mercado, D.; Sotomayor-Ramírez, D.; Cruz-Rodríguez, L. Microbial communities and enzymatic activities under different management in semiarid soils. Appl. Soil Ecol. 2008, 38, 249–260. [Google Scholar] [CrossRef]
- Furtak, K.; Gawryjołek, K.; Gajda, A.M.; Gałązka, A. Effects of maize and winter wheat grown in different cultivation techniques on biological activity of soil. Plant Soil Environ. 2017, 63, 449–454. [Google Scholar] [CrossRef]
- Gałązka, A.; Grządziel, J. Fungal Genetics and Functional Diversity of Microbial Communities in the Soil under Long-Term Monoculture of Maize Using Different Cultivation Techniques. Front. Microbiol. 2018, 9, 76. [Google Scholar] [CrossRef] [Green Version]
- Wolińska, A.; Frąc, M.; Oszust, K.; Szafranek-Nakonieczna, A.; Zielenkiewicz, U.; Stępniewska, Z. Microbial biodiversity of meadows under different modes of land use: Catabolic and genetic fingerprinting. World J. Microbiol. Biotechnol. 2017, 33, 154. [Google Scholar] [CrossRef]
- Jansson, J.K.; Hofmockel, K.S. The soil microbiome-from metagenomics to metaphenomics. Curr. Opin. Microbiol. 2018, 43, 162–168. [Google Scholar] [CrossRef]
- Torsvik, V.; Øvreås, L. Microbial diversity and function in soil: From genes to ecosystems. Curr. Opin. Microbiol. 2002, 5, 240–245. [Google Scholar] [CrossRef]
- Wagg, C.; Bender, S.F.; Widmer, F.; van der Heijden, M.G.A. Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc. Natl. Acad. Sci. USA 2014, 111, 5266–5270. [Google Scholar] [CrossRef] [Green Version]
- Delgado-Baquerizo, M.; Oliviero, A.M.; Brewer, T.E.; Benavent-González, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A global atlas of the dominant bacteria found in soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef] [Green Version]
- Girvan, M.S.; Billimore, J.; Pretty, J.N.; Osborn, A.M.; Ball, A.S. Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl. Environ. Microbiol. 2003, 69, 1800–1809. [Google Scholar] [CrossRef]
- Ma, F.; Wu, J.; Wang, L.; Yang, J.; Li, S.; Li, Z.; Zhag, X. Characterization of the microbial community in the rhizosphere of Phragmites australis (cav.) trin ex steudel growing in the Sun Island Wetland. Water Environ. Res. 2010, 86, 258–268. [Google Scholar] [CrossRef]
- Rousk, J.; Brookes, P.C.; Bååth, E. Contrasting soil pH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization. Appl. Environ. Microbiol. 2009, 75, 1589–1596. [Google Scholar] [CrossRef]
- Brochier-Armanet, C.; Boussau, B.; Gribaldo, S.; Forterre, P. Mesophilic crenarchaeota: Proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 2008, 6, 245–252. [Google Scholar] [CrossRef]
- Pester, M.; Schleper, C.; Wagner, M. The Thaumarchaeota: An emerging view of their phylogeny and ecophysiology. Curr. Opin. Microbiol. 2011, 14, 300–306. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.M.; Wang, N.; Hu, Y.B.; Sun, G.Y. Changes in soil physicochemical properties and soil bacterial community in mulberry (Morus alba L.)/alfalfa (Medicago sativa L.) intercropping system. Microbiol. Open 2018, 7, e00555. [Google Scholar] [CrossRef]
- Masse, J.; Prescott, C.E.; Renaut, S.; Terrat, Y.; Grayston, S.J. Plant community and nitrogen deposition as drivers of alpha and beta diversities of prokaryotes in reconstructed oil sand soils and natural boreal forest soils. Appl. Environ. Microbiol. 2017, 83, e03319-16. [Google Scholar] [CrossRef]
- Hansel, C.M.; Fendorf, S.; Jardine, P.M.; Francis, C.A. Changes in bacterial and archaeal community structure and functional diversity along a geochemically variable soil profile. Appl. Environ. Microbiol. 2008, 74, 1620–1633. [Google Scholar] [CrossRef]
- Wang, N.F.; Zhang, T.; Zhang, F.; Wang, E.T.; He, J.F.; Ding, H.; Zhang, B.T.; Liu, J.; Ran, X.B.; Zang, J.Y. Diversity and structure of soil bacterial communities in the Fildes Region (maritime Antarctica) as revealed by 454 pyrosequencing. Front. Microbiol. 2015, 6, 1188. [Google Scholar] [CrossRef] [Green Version]
- Laiz, L.; Miller, Z.; Jurado, V.; Akatova, E.; Sanchez-Moral, S.; Gonzalez, J.M.; Dionísio, A.; Macedo, M.F.; Saiz-Jimenez, C. Isolation of five Rubrobacter strains from biodeteriorated monuments. Naturwissenschaften 2009, 96, 71–79. [Google Scholar] [CrossRef]
- Norman, J.S.; King, G.M.; Friesen, M.L. Rubrobacter spartanus sp. nov., a moderately thermophilic oligotrophic bacterium isolated from volcanic soil. Int. J. Syst. Evol. MIcrobiol. 2017, 67, 3597–3602. [Google Scholar] [CrossRef]
- Kuske, C.R.; Yeager, C.M.; Johnson, S.; Ticknor, L.O.; Belnap, J. Response and resilience of soil biocrust bacterial communities to chronic physical disturbance in arid shrublands. ISME J. 2011, 6, 886–897. [Google Scholar] [CrossRef] [Green Version]
- Wolińska, A.; Stępniewska, Z.; Szymańska, E. Dehydrogenase activity of soil microorganisms and the total DNA level in soil of different use. J. Agric. Sci. Technol. B 2013, 3, 613–622. [Google Scholar]
- Park, J.E.; Lee, B.T.; Kim, B.Y.; Son, A. Bacterial community analysis of stabilized soils in proximity to an exhausted mine. Environ. Eng. Res. 2018, 23, 420–429. [Google Scholar] [CrossRef] [Green Version]
- Sheik, C.S.; Mitchell, T.W.; Rizvi, F.Z.; Rehman, Y.; Faisal, M.; Hasnain, S.; McInerney, M.J.; Krumholz, L.R. Exposure of soil microbial communities to chromium and arsenic alters their diversity and structure. PLoS ONE 2012, 7, e40059. [Google Scholar] [CrossRef]
- Hur, M.; Kim, Y.; Song, H.R.; Kim, J.M.; Choi, Y.I.; Yi, H. Effect of genetically modified poplars on soil microbial communities during the phytoremediation of waste mine tailings. Appl. Environ. Microbiol. 2011, 77, 7611–7619. [Google Scholar] [CrossRef]
- Flores-Rentería, D.; Rincón, A.; Valladares, F.; Yuste, J.C. Agricultural matrix affects differently the alpha and beta structural and functional diversity of soil microbial communities in a fragmented Mediterranean holm oak forest. Soil Biol. Biochem. 2016, 92, 79–90. [Google Scholar] [CrossRef]
- Gałązka, A.; Gawryjołek, K.; Grządziel, J.; Frąc, M.; Księżak, J. Microbial community diversity and the interaction of soil under maize growth in different Cultivation techniques. Plant Soil Environ. 2017, 63, 264–270. [Google Scholar] [CrossRef]
- Grządziel, J.; Furtak, K.; Gałązka, A. Community-level physiological profiles of microorganisms from different types of soil that are characteristic to Poland—A long-term Microplot experiment. Sustainability 2019, 11, 56. [Google Scholar] [CrossRef]
- Johns, C. Living Soils: The Role of Microorganisms in Soil Health. Strategic Analysis Paper. Future Directions International Pty Ltd. Available online: http://www.futuredirections.org.au/publication/ living-soils-role-microorganisms-soil-health/ (accessed on 23 June 2019).
- Li, G.; Kim, S.; Park, M.; Son, Y. Short-term effects of experimental warming and precipitation manipulation on soil microbial biomass C and N, community substrate utilization patterns and community composition. Pedosphere 2017, 27, 714–724. [Google Scholar] [CrossRef]
- Yan, N.; Marschner, P.; Cao, W.; Zuo, C.; Qiun, W. Influence of salinity and water content on soil microorganisms. Int. Soil Water Conserv. Res. 2015, 3, 316–323. [Google Scholar] [CrossRef] [Green Version]
- Bednarek, W.; Dresler, S.; Tkaczyk, P.; Hanaka, A. Physicochemical Properties of surface soil layer after the flood in the middle Vistula river valley. J. Elementol. 2014, 19, 17–29. [Google Scholar] [CrossRef]
- Lennon, J.T.; Jones, S.E. Microbial Seed Banks: The Ecological and Evolutionary Implications of Dormancy. Nat. Rev. Microbiol. 2011, 9, 119–130. [Google Scholar] [CrossRef]
- Ross, M.; Goberna, M.; Pascual, J.A.; Klammer, S.; Insam, H. 16S rDNA analysis reveals low microbial diversity in community level physiological profile assays. J. Microbiol. Methods 2008, 72, 221–226. [Google Scholar] [CrossRef]
- Griffiths, B.S.; Ritz, K.; Bardgett, R.D.; Cook, R.; Christensen, S.; Ekulund, F.; Sørensen, S.J.; Bååth, E.; Bloem, J.; de Ruiter, P.C.; et al. Ecosystem response of pasture soil communities to fumigation-induced microbial diversity reductions: An examination of the biodiversity-ecosystem function relationship. Oikos 2000, 90, 279–294. [Google Scholar] [CrossRef]
- Wertz, S.; Degrange, V.; Prosser, F.P.; Commeauc, C.; Guillaumaud, N.; Le Roux, X. Decline of soil microbial diversity does not influence the resistance and resilience of key soil microbial functional groups following a model disturbance. Environ. Microbiol. 2007, 9, 2211–2219. [Google Scholar] [CrossRef]





| Abbreviation | Location | GPS Coordinates | Distance from the Riverbed |
|---|---|---|---|
| F1 | Wojszyn, Puławy County | 51°20’03.4″ N 21°56’43.2″ E | 525 m |
| F2 | Janowiec (1), Puławy County | 51°19’29.9″ N 21°55’19.2″ E | 80 m |
| F3 | Janowiec (2), Puławy County | 51°19’14.4″ N 21°54’42.9″ E | 225 m |
| Index | Formula and Description | References |
|---|---|---|
| AWCD (average well color development) | [32] | |
| C—reading of the well OD590; | ||
| R—reading of the control well OD590; | ||
| n—number of substrates on an EcoPlate™ (n = 31). | ||
| Shannon diversity (H’) | [33] | |
| pi—proportional color development of the well over the total colour development of all wells (96) of a plate; | ||
| n—number of substrates on an EcoPlate™ (n = 31). | ||
| Shannon evenness (E) | [34] | |
| R—substrate utilization richness (the number of wells with colour development) | ||
| Richness (R) | The number of wells with color development—number of utilised substrates with OD590 ≥ 0.25 | [35] |
| Soil Sample | Soil Texture, mm (%) | Textural Classes | pHKCl | RDC (g 100g−1 soil) | TC (%) | OC (%) | TN (%) | OM (%) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2.0–0.05 | 0.05–0.002 | <0.002 | USDA 1 | PTG 2 | |||||||
| F1 | 58 | 38 | 4 | SL | gp | 6.96 | 0.16 a | 3.55 ± 0.02 a | 2.78± 0.04 a | 0.28± 0.05 a | 4.79± 0.06 a |
| F2 | 67 | 30 | 3 | SL | gp | 6.94 | 0.32 a | 2.07 ± 0.01 b | 1.72± 0.02 b | 0.17± 0.01 b | 2.96± 0.03 b |
| F3 | 92 | 8 | 0 | S | pl | 6.75 | n.d.* | 0.61 ± 0.01 c | 0.57± 0.02 c | 0.06± 0.01 c | 0.98± 0.04 c |
| Soil Sample | Metal Content (g kg−1 soil) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Al | Ba | Ca | Co | Cu | Cr | Fe | K | Li | Mg | Mn | Pb | Zn | |
| F1 | 37.28 ± 0.49 | 0.21 ± 0.00 | 15.25 ± 0.10 | 0.02 ± 0.00 | 0.05 ± 0.00 | 0.09 ± 0.00 | 32.77 ± 0.01 | 8.83 ± 0.06 | 0.04 ± 0.00 | 8.55 ± 0.01 | 1.35 ± 0.00 | 0.06 ± 0.00 | 0.48 ± 0.00 |
| F2 | 27.17 ± 0.07 | 0.15 ± 0.00 | 7.47 ± 0.01 | 0.01 ± 0.00 | 0.03 ± 0.00 | 0.06 ± 0.00 | 20.42 ± 0.42 | 7.17 ± 0.07 | 0.02 ± 0.00 | 5.37 ± 0.07 | 0.79 ± 0.02 | 0.03 ± 0.00 | 0.22 ± 0.00 |
| F3 | 8.75 ± 0.19 | 0.05 ± 0.00 | 1.36 ± 0.02 | 0.00 ± 0.00 | 0.01 ± 0.00 | 0.02 ± 0.00 | 6.58 ± 0.06 | 2.57 ± 0.02 | 0.01 ± 0.00 | 1.43 ± 0.01 | 0.23 ± 0.00 | 0.01 ± 0.00 | 0.04 ± 0.00 |
| Soil Sample | MBC (µgC g−1 d.m. of soil) | MBN (µgN g−1 d.m. of soil) | |
|---|---|---|---|
| F1 | 574.67 ± 13.3 a | 105.09 ± 4.5 a | |
| F2 | 191.11 ± 6.7 b | 32.95 ± 2.2 b | |
| F3 | 84.36 ± 9.7 c | 11.42 ± 1.2 c |
| Soil Sample | AWCD | H’ | E | R |
|---|---|---|---|---|
| F1 | 1.13 ± 0.08 b | 3.35 ± 0.02 a | 0.94 ± 0.01 a | 29.33 ± 0.58 a |
| F2 | 1.66 ± 0.11 a | 3.38 ± 0.02 a | 0.86 ± 0.01 b | 30.33 ± 0.58 a |
| F3 | 1.83 ± 0.06 a | 3.35 ± 0.02 a | 0.83 ± 0.00 c | 29.67 ± 0.58 a |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Furtak, K.; Grządziel, J.; Gałązka, A.; Niedźwiecki, J. Analysis of Soil Properties, Bacterial Community Composition, and Metabolic Diversity in Fluvisols of a Floodplain Area. Sustainability 2019, 11, 3929. https://doi.org/10.3390/su11143929
Furtak K, Grządziel J, Gałązka A, Niedźwiecki J. Analysis of Soil Properties, Bacterial Community Composition, and Metabolic Diversity in Fluvisols of a Floodplain Area. Sustainability. 2019; 11(14):3929. https://doi.org/10.3390/su11143929
Chicago/Turabian StyleFurtak, Karolina, Jarosław Grządziel, Anna Gałązka, and Jacek Niedźwiecki. 2019. "Analysis of Soil Properties, Bacterial Community Composition, and Metabolic Diversity in Fluvisols of a Floodplain Area" Sustainability 11, no. 14: 3929. https://doi.org/10.3390/su11143929






