Adenovirus Vector-Derived VA-RNA-Mediated Innate Immune Responses
Abstract
: The major limitation of the clinical use of replication-incompetent adenovirus (Ad) vectors is the interference by innate immune responses, including induction of inflammatory cytokines and interferons (IFN), following in vivo application of Ad vectors. Ad vector-induced production of inflammatory cytokines and IFNs also results in severe organ damage and efficient induction of acquired immune responses against Ad proteins and transgene products. Ad vector-induced innate immune responses are triggered by the recognition of Ad components by pattern recognition receptors (PRRs). In order to reduce the side effects by Ad vector-induced innate immune responses and to develop safer Ad vectors, it is crucial to clarify which PRRs and which Ad components are involved in Ad vector-induced innate immune responses. Our group previously demonstrated that myeloid differentiating factor 88 (MyD88) and toll-like receptor 9 (TLR9) play crucial roles in the Ad vector-induced inflammatory cytokine production in mouse bone marrow-derived dendritic cells. Furthermore, our group recently found that virus associated-RNAs (VA-RNAs), which are about 160 nucleotide-long non-coding small RNAs encoded in the Ad genome, are involved in IFN production through the IFN-β promoter stimulator-1 (IPS-1)-mediated signaling pathway following Ad vector transduction. The aim of this review is to highlight the Ad vector-induced innate immune responses following transduction, especially VA-RNA-mediated innate immune responses. Our findings on the mechanism of Ad vector-induced innate immune responses should make an important contribution to the development of safer Ad vectors, such as an Ad vector lacking expression of VA-RNAs.1. Introduction
The first-generation E1-deleted adenovirus (Ad) vector (FG-Ad vector) is one of the most promising vectors for gene therapy, as well as basic research due to its advantages as a gene delivery vehicle. FG-Ad vectors are relatively easy to construct, can be produced at a high titer, and have a high transduction efficiency into a wide spectrum of dividing and non-dividing cells in vitro and in vivo. However, application of FG-Ad vectors has been limited to local administration in most clinical trials of gene therapy, since the systemic administration of Ad vectors induces both adaptive and innate immune responses with its humoral and cell-mediated components [1,2].
In the case of adaptive immune response, capsid antigens are largely responsible for the specific immunity toward Ad vectors. In addition, in the FG-Ad vector, leaky expression of viral genes from the vector stimulates an immune response against Ad vector-transduced cells [3,4]. In a replication-incompetent FG-Ad vector, the E1A gene, which encodes an essential transactivator of the other viral genes, is deleted, in addition to the E1B gene. Therefore, theoretically, no other viral genes should be expressed following transduction; however, viral genes from the vector genome, including the E2A and E4 genes, are indeed expressed, which leads to an induction of cellular immunity against Ad proteins as well as to Ad protein-induced toxicity [5-11]. Such Ad protein-induced cellular immunity and toxicity frequently cause both an elimination of Ad vector-transduced cells and tissue damages, leading to short-lived transgene expression [5-7,9,10,12,13]. We recently quantitatively analyzed the leaky expression profiles of Ad genes using real-time RT-PCR following transduction with FG-Ad vectors [14]. The pIX and E4 genes were most highly expressed among the Ad genes examined, and their expression levels were 1/50 to 1/5000 of the expression levels observed with the same MOI of the wild-type Ad. The cytotoxic T lymphocyte (CTL) response can be elicited against these viral gene products and/or transgene products expressed in the transduced cells. On the other hand, the expression levels of E2B and major capsid proteins (including the hexon, penton base, and fiber proteins) were almost the same or even slightly above those observed in mock-transduced cells [14].
To reduce cell-mediated immune responses against viral gene products expressed in the transduced cells, “helper-dependent (HD)” or “gutted” Ad vectors have been developed. In this vector, all viral genes are deleted except the inverted terminal repeat (ITR) sequences at both ends and the packaging signal. The deletion of all viral protein-coding regions from the Ad genome improves the prospects of Ad vectors for long-term gene expression, suggesting that the immunogenic toxicities induced by HD-Ad vectors are greatly reduced [15].
Humoral virus-neutralizing antibody responses against the Ad capsid itself are another limitation, preventing transduction upon the subsequent administration of vectors of the same serotype. Because hexons are primarily targeted by neutralizing antibodies, hexon modification has been reported to allow for escape from neutralizing antibodies [16]. As other strategies, Ad vectors belonging to subgroups other than Ad serotype 5, such as Ad serotype 11 or 35, or to species other than humans, have also been developed [17-21].
Compared with the adaptive immunity to Ad vectors, the mechanism of Ad vector-induced innate immune responses is less understood; however, several studies have reported which cellular molecules are involved in and which components of Ad trigger the Ad vector-induced innate immune responses [22-25]. In addition, we demonstrated that virus associated RNA (VA-RNA) I and II, which are about 160 nucleotide-long non-coding RNAs encoded in the Ad genome (bp 10620-10779 and 10876-11038), are transcribed from a conventional FG-Ad vector as well as the wild-type Ad, and that VA-RNAs trigger innate immune responses through the IFN-β promoter stimulator-1 (IPS-1) [23]. In this report, we review the Ad vector-mediated innate immune responses, especially VA-RNA-mediated innate immune responses.
2. Ad vector-Mediated Innate Immune Responses
2.1. Biodistribution of intravenously injected Ad vectors and production of cytokines/chemokines
Systemically administered Ad vectors are rapidly cleared from the blood of mice, with a half-life of less than 3 min [26-28]. Liver Kupffer cells play a central role in clearing the Ad genome from the bloodstream [29-31]. It has been proposed that a low dose of Ad vectors (∼1010 vector particles) is rapidly sequestered by Kupffer cells (liver macrophages), while higher doses of Ad vectors are delivered into both Kupffer cells and hepatocytes, leading to a nonlinear dose response in hepatic transgene expression [32]. At a dose of 3.0 × 1010 vector particles per mouse, Ad vectors are likely to be equally distributed to the Kupffer and hepatocytes [27,33]. Despite the high uptake of Ad vectors by Kupffer cells, the Ad vector-mediated transduction efficiencies in Kupffer cells were much lower than those in the hepatocytes, indicating the uptake of Ad vectors by Kupffer cells is a function of phagocytosis rather than a receptor-mediated infectious pathway [28]. The spleen is the second organ that intravenously injected Ad vectors accumulate in. Since liver and spleen contain many immune cells, including liver Kupffer cells, splenic dendritic cells (DCs) and macrophages, these cells had been assumed to be responsible for the production of inflammatory cytokines/chemokines such as interleukin (IL)-6, tumor necrosis factor (TNF)-α, and Interferon gamma-induced protein 10 kDa (IP-10), and regulated on activation, normal T cell expressed and secreted (RANTES). The production of these inflammatory cytokines/chemokines causes the activation of an innate immune response [34,35]. Recent reports suggested that the spleen is the major site of cytokine, chemokine, and interferon (IFN) production. When the mice are splenectomized, IL-6 production is decreased upon Ad vector injection [36]. Reverse transcriptase-polymerase chain reaction (RT-PCR) analysis of the liver and spleen after systemic Ad vector injection suggests that IL-6 as well as other immune cytokines, chemokines, and IFNs were mainly produced from the spleen, especially from conventional dendritic cells (DC) (CD11c+B220− cells), not the liver [37]. The fiber-modified Ad vector containing a stretch of lysine residues (K7 (KKKKKKK) peptide), which target heparan sulfates on the cellular surface, greatly reduced IL-6 production after intravenous injection into mice, possibly due to the reduced biodistribution of the Ad vector to the spleen [37].
Excessive complement activation has also been reported to be involved in Ad vector-mediated innate immune responses [38,39]. Furthermore, Xu et al. reported that Ad vectors are opsonized by immunoglobulin M antibodies and complement, leading to clearance of Ad vectors by Kupffer cells, which is dependent on scavenger receptors [40]. Blood coagulation factor X binds to hypervariable regions of the Ad5 hexon, leading to liver transduction and hepatotoxicity [41-45]. Modification of the hypervariable regions of hexon resulted in decreases of Ad vector-transduction of hepatocytes with potential evasion from Kupffer cells [46]. The interaction of Ad vectors with blood factors and the subsequent uptake by Kupffer cells might also play a role in the induction of innate immune responses.
2.2. Signaling pathway leading to innate immune responses
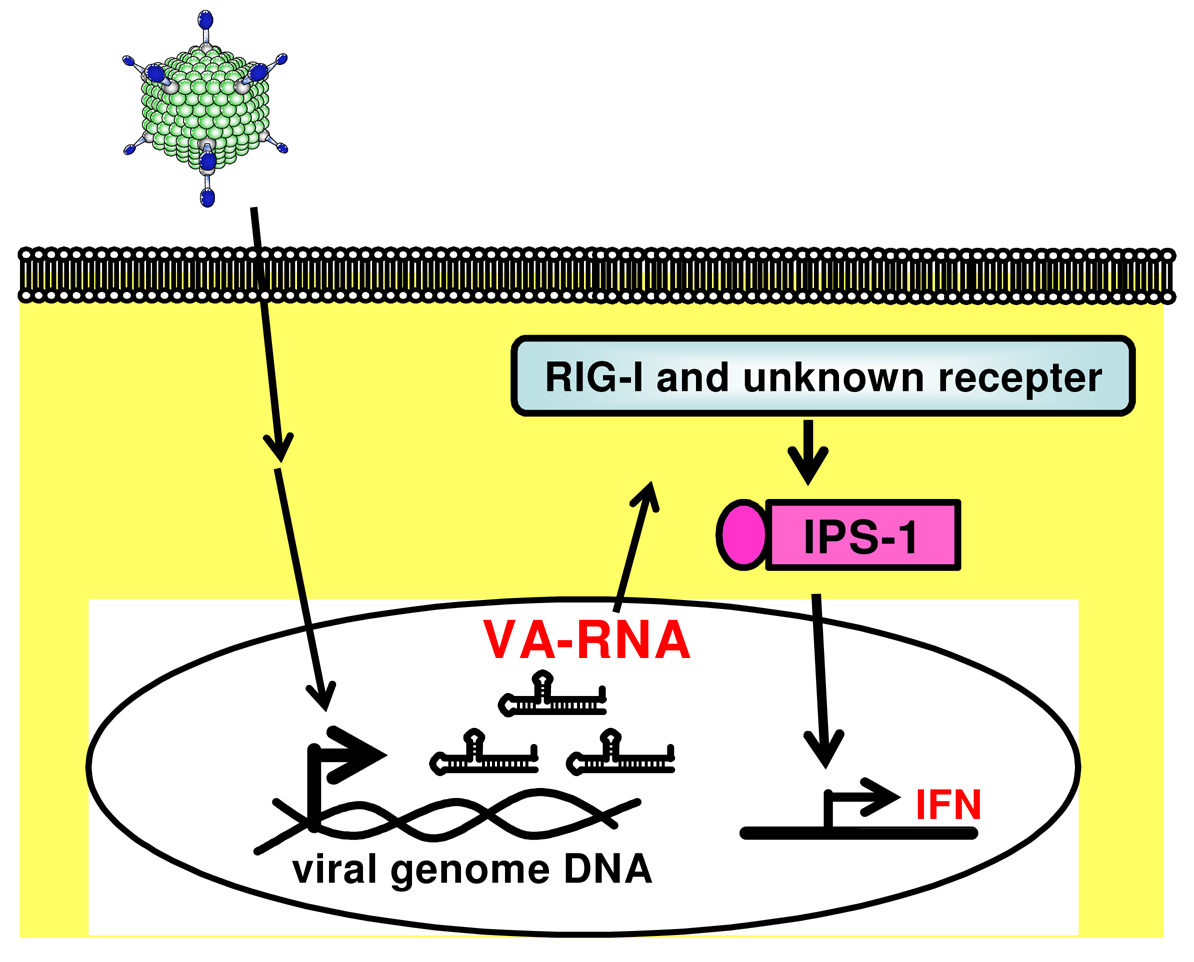
The possible components of FG-Ad vectors responsible for activating innate immune responses are capsid proteins, the viral genome (DNA), and viral transcripts. Induction of the antiviral innate immune response depends on the recognition of viral components by host pattern recognition receptors (PRRs). The most well-known PRRs are the Toll-like receptors (TLRs) that recognize pathogen-associated molecular patterns (PAMPs), including lipopolysaccharides, double-stranded RNA (dsRNA), single-strand RNA (ssRNA), and unmethylated CpG DNA [47]. After the recognition of PAMPs by TLRs, all of the TLRs, with the exception of TLR3, transduce intracellular signaling through the adaptor protein myeloid differentiating factor 88 (MyD88), which initiates a signaling cascade leading to the activation of NF-κB and IFN regulatory factors (IRFs). We and others have reported that Ad vectors elicit the production of inflammatory cytokines such as IL-6 and IL-12 in a TLR9-dependent manner in conventional dendritic cells (cDCs) [24,48]. Zhu et al. [49] demonstrated that the Ad vector-induced production of type I IFN by cDCs is mediated by a TLR9-independent pathway, whereas the Ad vector-induced production of type I IFN by plasmacytoid DCs is mediated by a TLR9-dependent pathway. The signaling pathway for type I IFN production after Ad vector treatment is known to differ from that for inflammatory cytokine production. In another PRR pathway, cytosolic RNAs are recognized by retinoic acid-inducible gene I (RIG-I)-like receptors, which include RIG-I and melanoma differentiation-associated gene 5 (Mda5) [50-52]. RIG-I and Mda5 contain RNA helicase domains that recognize viral dsRNA [53,54]. In addition, RIG-I recognizes ssRNA containing 5′-triphosphate [55-57]. RIG-I and Mda5 also contain N-terminal tandem caspase activation and recruitment domains (CARDs), which interact with the CARD domain of IFN-β promoter stimulator-1 (IPS-1, also known as MAVS, VISA, and Cardif) [58]. This interaction finally activates several transcriptional factors (e.g., NF-κB, IRF3, and IRF7) and induces the production of inflammatory cytokines and type I IFN.
3. VA-RNA-Mediated Innate Immune Response and an Ad Vector Lacking the Expression of VA-RNAs
3.1. Function of VA-RNAs and VA-RNA-mediated innate immune response
The Ad genome encodes two non-coding small RNAs, VA-RNA I (a major species) and VA-RNA II (a minor species), which are about 160 nucleotide-long non-coding RNAs encoded in the Ad serotype 5 genome regions, respectively, and which are transcribed by RNA polymerase III [59]. VA-RNAs have two internal transcription control elements (box A and box B), which in the case of VA-RNA I derived from Ad serotype 5 are located at bp 10629-10637 and bp 10673-10688, respectively [60,61]. Sequences of VA-RNAs vary considerably among Ad vectors from different serotypes; however, structural characteristics (i.e., the apical stem, the central domain, and the terminal domain) are conserved in all VA-RNAs and are structurally highly stable (Figure 1) [62,63]. The apical stem and the central domain are involved in Ad amplification through the association with double-stranded RNA-dependent protein kinase (PKR), as described below. The apical stem is essential for the association with PKR [64-67] and the central domain is required for efficient inhibition of PKR activation [68-71]. The terminal stem is recognized by Exportin-5, which is essential for nuclear export of VA-RNAs [72]. Furthermore, the terminal stem may be required for the stabilization of other domains and the protection from exonuclease activity in vivo [63]. VA-RNA I is rapidly synthesized and accumulated to very high levels, 108 molecules per cell, while VA-RNA II is synthesized at 20-fold lower levels, 5 × 106 molecules per cell, during the late phase of infection [59]. VA-RNAs are transcribed from a conventional FG-Ad vector as well as the wild-type Ad, and this transcription depends on RNA polymerase III [23].
In previous studies, VA-RNA I-deleted Ad grew 10-fold less well, while VA-RNA II-deleted Ad grew as well as the wild-type Ad; however, both VA-RNA I- and II-deleted Ad grew 60-fold less well (note that these VA-RNA mutant Ads contain the E1 gene) [73-77]. These results suggest that VA-RNA I is required for efficient translation of viral mRNAs and allows the efficient amplification of Ad, and that VA-RNA II also supports the replication, even though the effect is rather weak compared to that of VA-RNA I. The molecular mechanism of VA-RNAs in supporting the Ad amplification is, at least in part, due to antagonization of the antiviral action associated with the activation of PKR [77]. Following virus infection, dsRNA produced during virus replication can bind to two PKR molecules, leading to autophosphorylation of PKR itself and the subsequent activation of PKR. Activated PKR phosphorylates αsubunit of eukaryotic translation initiation factor-2 (eIF-2) (eIF-2α) [78], leading to the tight association between phosphorylated eIF-2α and eIF-2B [79]. eIF-2B is a guanosine nucleotide exchange factor and plays a crucial role on the conversion of GDP to GTP; however, the association between phosphorylated eIF-2α and eIF-2B captures eIF-2B and prevents the conversion of GDP on non-phosphorylated eIF-2α, resulting in inhibition of the translation of viral mRNA and/or to cell apoptosis [59,78,80-82]. Upon infection with an Ad, VA-RNA I is transcribed from the Ad genome in the infected cells. Subsequently, the VA-RNA I binds to PKR with high affinity, blocks the PKR activation, and enhances viral amplification [75,77,83]. Previous studies using mutant VA-RNAs have demonstrated that the apical stem and the central domain play an essential roles in the association between PKR and VA-RNA I [63,84,85]. In contrast, VA-RNA II only shows limited ability to block the activation of PKR [86]. Furthermore, not all Ad serotypes have VA-RNA II [63]. These suggest that VA-RNA II may not be essential in the Ad amplification.
Recently, several studies have demonstrated that VA-RNAs are also processed in a manner similar to microRNAs (miRNAs) [72,75,87-91]. VA-RNAs are transported to cytoplasm by interaction between those terminal stem and Exportin-5, and are then processed by Dicer. After being processed by Dicer, mivaRNAs are incorporated into the RNA-induced silencing complex (RISC), resulting in production of VA-RNA-derived miRNAs (mivaRNAs). In this processing step, VA-RNAs saturate Exportin-5, Dicer, and RISC, leading to the repression of endogenous miRNA-mediated regulation [88-90]. Surprisingly, approximately 80% of RISC immunopurified from cells transduced with the wild-type Ad is associated with mivaRNAs [89]. Both VA-RNA I and II are processed into mivaRNAs. mivaRNAI-137 and -138 are produced from VA-RNA I (Figure 1); however, only about 5% of total VA-RNA I is processed, because VA-RNAs have larger stem than miRNAs and this processing is not efficient [88-90]. On the other hand, VA-RNA II is mainly processed into mivaRNAII-138 derived from the 3′-strand of VA-RNA II [89,92]. Higher amounts of mivaRNAs derived from VA-RNA II were found in RISC, compared with mivaRNAs derived from VA-RNA I [89]. RISC containing mivaRNAs would suppress expression of target genes, developing cellular environment which is appropriate for virus amplification. Recently, Aparicio et al. reported that one of target genes for mivaRNAI-138 is TIA-1, which promotes the expression of proapoptotic proteins [91]. Several genes that are involved in the antiviral action would be suppressed by VA-RNAs in a manner similar to miRNA to support the amplification of Ads.
VA-RNA I is classified into two species, VA-RNA I (A) and VA-RNA I (G), according to the heterogeneity at the 5′-ends derived from the diversity of initiation of transcription [92-94]. VA-RNA I (G) is transcribed 3 nucleotides downstream of the transcription initiation site of VA-RNA I (A), is preferentially synthesized and accounts for about 75% of the total VA-RNA I [92,93]. VA-RNA I (A) and (G) are processed at the same position of the 3′-strand, generating mivaRNAI-137 and 138 (Figure 1) [89]. The 3′strand mivaRNAI is produced at much higher levels compared to the 5′-strand mivaRNAI (A) and (G); however, RISC assembled on the 5′strand mivaRNAI (A) shows higher cleavage activity than that assembled on the 3′strand mivaRNAI [92]. Further examination is required to clarify whether the heterogeneity of the 5′-end of VA-RNA I leads to the differences in mivaRNA-mediated regulation of gene expression.
Many viruses express some noncoding RNAs which probably control host cellular responses and maximize their replication in the infected host [95,96]. Among the best-characterized viral small RNAs are the Epstein-Barr virus (EBV)-encoded small RNAs (EBERs). EBERs are nonpolyadenylated, untranslated RNAs, transcribed by RNA polymerase III abundantly in latently EBV-infected cells, and form a stem-loop structure. The stem-loop structure of EBERs, which gives rise to dsRNA-like molecules, is recognized by RIG-I and induces expression of type I IFN and IFN-stimulated genes (ISGs) [97]. A recent report by Iwakiri et al. demonstrated that EBERs released from EBV-infected cells are recognized by TLR3, and induce type I IFN and proinflammatory cytokines in surrounding noninfected cells as well as in the EBV-infected cells [98]. These evidences suggest that virus-encoded small RNAs are key to not only inducing the innate immune response upon virus infection but also potentially enhancing the following adaptive immunity. We recently demonstrated that VA-RNAs induce the production of type I IFN (IFN-α and IFN-β), but they do not induce the production of inflammatory cytokines (IL-6 and IL-12), in mouse embryonic fibroblasts (MEFs) and granulocyte-macrophage colony-stimulating factor-generated bone marrow-derived dendritic cells (GM-DCs) (Figure 2) [23]. We also showed that IPS-1 is involved in VA-RNA-dependent IFN-β production in MEFs and is partially involved in type I IFN production in GM-DCs (Figure 2) [23]. Moreover, Minamitani et al. also reported that VA-RNAs induce the production of type I IFN through an RIG-I pathway [25]. A certain fraction of VA-RNA I possesses triphosphated 5′-termini [94]. Since ssRNA containing 5′-triphosphate activates RIG-I [55,99], 5′-triphosphate of VA-RNA I may be involved in RIG-I-dependent IFN induction. Furthermore, as described above, VA-RNA I is classified into two species, VA-RNA I (A) and VA-RNA I (G) [92-94]. This heterogeneity at 5′-termini of VA-RNA I would lead to the difference in the phosphorylation pattern at 5′-termini. Xu et al. demonstrated that higher amounts of mivaRNAI (G) possess the multi-phosphate at 5′-termini, compared with mivaRNAI (A). mivaRNAI (G) might induce type I IFN production more strongly than mivaRNAI (A) in a RIG-I-dependent manner; however, in our analysis using RIG-I-deficient mice, the production of type I IFN is independent of an RIG-I pathway [23].
3.2. Development of an Ad vector lacking the expression of VA-RNAs
The findings described above strongly suggest that a VA-RNA-deleted Ad (AdΔVR) vector could be less able to activate the innate immune response, providing a safer alternative to the FG-Ad vectors. Furthermore, development of the AdΔVR vector could provide important insights into unknown functions of VA-RNAs and lead to the discovery of other target genes of mivaRNAs derived from VA-RNAs.
We have developed an AdΔVR vector from which the transcriptional control elements of the VA-RNA expression were deleted. Some previous reports suggested that 293T cells expressing the SV40 large-T antigen or a PKR inhibitor (2-aminopurine, 2-AP) might support the replication and propagation of the AdΔVR vector as follows: the SV40 large-T antigen which antagonizes the translational inhibitory effect resulting from the activation of PKR in SV40-infected cells resulted in counteraction of cellular defense against virus infection [100]; 2-aminopurine (2-AP) interferes with the gene induction normally triggered by IFN, and supports the amplification of a VA-RNA-deleted mutant Ad (Sub720) [78,101]. So far, unfortunately, we have failed to generate an AdΔR vector by these approaches, probably due to the lower capability of propagating Ad vectors in 293T cells and cytotoxicity of 2-AP against HEK293 cells, respectively.
We prepared VA-RNA I-expressing HEK293 cells (VR293 cells) (Figure 3). VR293 cells allowed the generation and propagation of an AdΔVR vector without the need for any complicated processes or other supporting materials, such as a helper virus, although the recovered titer of the AdΔVR vector was low. The resulting AdΔVR vector showed a high-purity, no expression of VA-RNAs, and an efficient transduction capability [102]. Further studies are needed to develop an improved method for more efficient propagation of AdΔVR vectors (e.g., generation of an improved cell line expressing VA-RNA II as well as VA-RNA I; the use of other PKR inhibitors and IFN-inhibitors with low cytotoxicity). The induction of both the early innate and following adaptive immune responses by the AdΔVR vector should be evaluated in vitro and in vivo (note that more efficient propagation of AdΔVR vectors is required to analyze the immune response by AdΔVR vectors). The AdΔVR vector could be one of the most variable tools for the analysis of VA-RNA-mediated innate immune response following transduction with Ad vectors.
4. Conclusions
In the present paper, we reviewed Ad vector-mediated immunogenic toxicities, especially innate immune responses and VA-RNA. Ad vectors activate innate immune responses through the capsid protein, the viral genome (DNA), and/or viral transcripts. VA-RNAs produced in the Ad vector-transduced cells also stimulate the innate immune responses. It is essential to clarify the mechanism of the innate immune responses triggered by the systemic administration of Ad vectors in order to achieve a safe method of gene therapy using Ad vectors. In addition, such an improved understanding of Ad vector-induced innate immune responses is essential for the successful gene therapy as well as the development of safe vectors.


Acknowledgments
This work was supported by grants from the Ministry of Health, Labour, and Welfare of Japan, and a Grant-in-Aid for Scientific Research (B) of the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan.
We thank Ms. Ong Tyng Tyng for critical reading of the manuscript.
References
- Sakurai, H.; Kawabata, K.; Sakurai, F.; Nakagawa, S.; Mizuguchi, H. Innate immune response induced by gene delivery vectors. Int. J. Pharm. 2008, 354, 9–15. [Google Scholar]
- Xu, Z.L.; Mizuguchi, H.; Sakurai, F.; Koizumi, N.; Hosono, T.; Kawabata, K.; Watanabe, Y.; Yamaguchi, T.; Hayakawa, T. Approaches to improving the kinetics of adenovirus-delivered genes and gene products. Adv. Drug Deliv. Rev. 2005, 57, 781–802. [Google Scholar]
- Yang, Y.; Ertl, H.C.; Wilson, J.M. MHC class I-restricted cytotoxic T lymphocytes to viral antigens destroy hepatocytes in mice infected with E1-deleted recombinant adenoviruses. Immunity 1994, 1, 433–442. [Google Scholar]
- Yang, Y.; Li, Q.; Ertl, H.C.; Wilson, J.M. Cellular and humoral immune responses to viral antigens create barriers to lung-directed gene therapy with recombinant adenoviruses. J. Virol. 1995, 69, 2004–2015. [Google Scholar]
- Yang, Y.; Nunes, F.A.; Berencsi, K.; Furth, E.E.; Gönczöl, E.; Wilson, J.M. Cellular immunity to viral antigens limits E1-deleted adenoviruses for gene therapy. Proc. Natl. Acad. Sci. USA 1994, 91, 4407–4411. [Google Scholar]
- Yang, Y.; Ertl, H.C.; Wilson, J.M. MHC class I-restricted cytotoxic T lymphocytes to viral antigens destroy hepatocytes in mice infected with E1-deleted recombinant adenoviruses. Immunity 1994, 1, 433–442. [Google Scholar]
- Yang, Y.; Xiang, Z.; Ertl, H.C.; Wilson, J.M. Upregulation of class I major histocompatibility complex antigens by interferon gamma is necessary for T-cell-mediated elimination of recombinant adenovirus-infected hepatocytes in vivo. Proc. Natl. Acad. Sci. USA 1995, 92, 7257–7261. [Google Scholar]
- Gorziglia, M.I.; Kadan, M.J.; Yei, S.; Lim, J.; Lee, G.M.; Luthra, R.; Trapnell, B.C. Elimination of both E1 and E2 from adenovirus vectors further improves prospects for in vivo human gene therapy. J. Virol. 1996, 70, 4173–4178. [Google Scholar]
- Gao, G.P.; Yang, Y.; Wilson, J.M. Biology of adenovirus vectors with E1 and E4 deletions for liver-directed gene therapy. J. Virol. 1996, 70, 8934–8943. [Google Scholar]
- Lusky, M.; Christ, M.; Rittner, K.; Dieterle, A.; Dreyer, D.; Mourot, B.; Schultz, H.; Stoeckel, F.; Pavirani, A.; Mehtali, M. In vitro and in vivo biology of recombinant adenovirus vectors with E1, E1/E2A, or E1/E4 deleted. J. Virol. 1998, 72, 2022–2032. [Google Scholar]
- Sharma, A.; Bangari, D.S.; Tandon, M.; Pandey, A.; HogenEsch, H.; Mittal, S.K. Comparative analysis of vector biodistribution, persistence and gene expression following intravenous delivery of bovine, porcine and human adenoviral vectors in a mouse model. Virology 2009, 386, 44–54. [Google Scholar]
- Wang, Q.; Finer, M.H. Second-generation adenovirus vectors. Nat. Med. 1996, 2, 714–716. [Google Scholar]
- Chen, D.; Murphy, B.; Sung, R.; Bromberg, J.S. Adaptive and innate immune responses to gene transfer vectors: role of cytokines and chemokines in vector function. Gene Therapy 2003, 10, 991–998. [Google Scholar]
- Shimizu, K.; Sakurai, F.; Machitani, M.; Katayama, K.; Mizuguchi, H. Quantitative analysis of the leaky expression of adenovirus genes in cells transduced with a replication-incompetent adenovirus vector. Mol. Pharm. 2011. In Press. [Google Scholar]
- Palmer, D.J.; Ng, P. Helper-dependent adenoviral vectors for gene therapy. Hum. Gene Ther. 2005, 16, 1–16. [Google Scholar]
- Roberts, D.M.; Nanda, A.; Havenga, M.J.; Abbink, P.; Lynch, D.M.; Ewald, B.A.; Liu, J.; Thorner, A.R.; Swanson, P.E.; Gorgone, D.A.; et al. Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature 2006, 441, 239–243. [Google Scholar]
- Farina, S.F.; Gao, G.P.; Xiang, Z.Q.; Rux, J.J.; Burnett, R.M.; Alvira, M.R.; Marsh, J.; Ertl, H.C.; Wilson, J.M. Replication-defective vector based on a chimpanzee adenovirus. J. Virol. 2001, 75, 11603–11613. [Google Scholar]
- Sakurai, F.; Mizuguchi, H.; Hayakawa, T. Efficient gene transfer into human CD34+ cells by an adenovirus type 35 vector. Gene Therapy 2003, 10, 1041–1048. [Google Scholar]
- Vogels, R.; Zuijdgeest, D.; van Rijnsoever, R.; Hartkoorn, E.; Damen, I.; de Bethune, M.P.; Kostense, S.; Penders, G.; Helmus, N.; Koudstaal, W.; et al. Replication-deficient human adenovirus type 35 vectors for gene transfer and vaccination: efficient human cell infection and bypass of preexisting adenovirus immunity. J. Virol. 2003, 77, 8263–8271. [Google Scholar]
- Holterman, L.; Vogels, R.; van der Vlugt, R.; Sieuwerts, M.; Grimbergen, J.; Kaspers, J.; Geelen, E.; van der Helm, E.; Lemckert, A.; Gillissen, G.; et al. Novel replication-incompetent vector derived from adenovirus type 11 (Ad11) for vaccination and gene therapy: low seroprevalence and non-cross-reactivity with Ad5. J. Virol. 2004, 78, 13207–13215. [Google Scholar]
- Seshidhar Reddy, P.; Ganesh, S.; Limbach, M.P.; Brann, T.; Pinkstaff, A.; Kaloss, M.; Kaleko, M.; Connelly, S. Development of adenovirus serotype 35 as a gene transfer vector. Virology 2003, 311, 384–393. [Google Scholar]
- Muruve, D.A.; Cotter, M.J.; Zaiss, A.K.; White, L.R.; Liu, Q.; Chan, T.; Clark, S.A.; Ross, P.J.; Meulenbroek, R.A.; Maelandsmo, G.M.; et al. Helper-dependent adenovirus vectors elicit intact innate but attenuated adaptive host immune responses in vivo. J. Virol. 2004, 78, 5966–5972. [Google Scholar]
- Yamaguchi, T.; Kawabata, K.; Kouyama, E.; Ishii, K.J.; Katayama, K.; Suzuki, T.; Kurachi, S.; Sakurai, F.; Akira, S.; Mizuguchi, H. Induction of type I interferon by adenovirus-encoded small RNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 17286–17291. [Google Scholar]
- Yamaguchi, T.; Kawabata, K.; Koizumi, N.; Sakurai, F.; Nakashima, K.; Sakurai, H.; Sasaki, T.; Okada, N.; Yamanishi, K.; Mizuguchi, H. Role of MyD88 and TLR9 in the innate immune response elicited by serotype 5 adenoviral vectors. Hum. Gene Ther. 2007, 18, 753–762. [Google Scholar]
- Minamitani, T.; Iwakiri, D.; Takada, K. Adenovirus virus-associated RNAs induce type I interferon expression through a RIG-I-mediated pathway. J. Virol. 2011, 85, 4035–4040. [Google Scholar]
- Alemany, R.; Suzuki, K.; Curiel, D.T. Blood clearance rates of adenovirus type 5 in mice. J. Gen. Virol. 2000, 81, 2605–2609. [Google Scholar]
- Koizumi, N.; Mizuguchi, H.; Sakurai, F.; Yamaguchi, T.; Watanabe, Y.; Hayakawa, T. Reduction of natural adenovirus tropism to mouse liver by fiber-shaft exchange in combination with both CAR- and alphav integrin-binding ablation. J. Virol. 2003, 77, 13062–13072. [Google Scholar]
- Sakurai, F.; Mizuguchi, H.; Yamaguchi, T.; Hayakawa, T. Characterization of in vitro and in vivo gene transfer properties of adenovirus serotype 35 vector. Mol. Ther. 2003, 8, 813–821. [Google Scholar]
- Lieber, A.; He, C.Y.; Meuse, L.; Schowalter, D.; Kirillova, I.; Winther, B.; Kay, M.A. The role of Kupffer cell activation and viral gene expression in early liver toxicity after infusion of recombinant adenovirus vectors. J. Virol. 1997, 71, 8798–8807. [Google Scholar]
- Wolff, G.; Worgall, S.; van Rooijen, N.; Song, W.R.; Harvey, B.G.; Crystal, R.G. Enhancement of in vivo adenovirus-mediated gene transfer and expression by prior depletion of tissue macrophages in the target organ. J. Virol. 1997, 71, 624–629. [Google Scholar]
- Worgall, S.; Wolff, G.; Falck-Pedersen, E.; Crystal, R.G. Innate immune mechanisms dominate elimination of adenoviral vectors following in vivo administration. Hum. Gene Ther. 1997, 8, 37–44. [Google Scholar]
- Tao, N.; Gao, G.P.; Parr, M.; Johnston, J.; Baradet, T.; Wilson, J.M.; Barsoum, J.; Fawell, S.E. Sequestration of adenoviral vector by Kupffer cells leads to a nonlinear dose response of transduction in liver. Mol. Ther. 2001, 3, 28–35. [Google Scholar]
- Koizumi, N.; Kawabata, K.; Sakurai, F.; Watanabe, Y.; Hayakawa, T.; Mizuguchi, H. Modified adenoviral vectors ablated for coxsackievirus-adenovirus receptor, alphav integrin, and heparan sulfate binding reduce in vivo tissue transduction and toxicity. Hum. Gene Ther. 2006, 17, 264–279. [Google Scholar]
- Liu, Q.; Muruve, D.A. Molecular basis of the inflammatory response to adenovirus vectors. Gene Therapy 2003, 10, 935–940. [Google Scholar]
- Zhang, Y.; Chirmule, N.; Gao, G.P.; Qian, R.; Croyle, M.; Joshi, B.; Tazelaar, J.; Wilson, J.M. Acute cytokine response to systemic adenoviral vectors in mice is mediated by dendritic cells and macrophages. Mol. Ther. 2001, 3, 697–707. [Google Scholar]
- De Geest, B.; Snoeys, J.; Van Linthout, S.; Lievens, J.; Collen, D. Elimination of innate immune responses and liver inflammation by PEGylation of adenoviral vectors and methylprednisolone. Hum. Gene Ther. 2005, 16, 1439–1451. [Google Scholar]
- Koizumi, N.; Yamaguchi, T.; Kawabata, K.; Sakurai, F.; Sasaki, T.; Watanabe, Y.; Hayakawa, T.; Mizuguchi, H. Fiber-modified adenovirus vectors decrease liver toxicity through reduced interleukin 6 production. J. Immunol. 2007, 178, 1767–1773. [Google Scholar]
- Kiang, A.; Hartman, Z.C.; Everett, R.S.; Serra, D.; Jiang, H.; Frank, M.M.; Amalfitano, A. Multiple innate inflammatory responses induced after systemic adenovirus vector delivery depend on a functional complement system. Mol. Ther. 2006, 14, 588–598. [Google Scholar]
- Tian, J.; Xu, Z.; Smith, J.S.; Hofherr, S.E.; Barry, M.A.; Byrnes, A.P. Adenovirus activates complement by distinctly different mechanisms in vitro and in vivo: indirect complement activation by virions in vivo. J. Virol. 2009, 83, 5648–5658. [Google Scholar]
- Xu, Z.; Tian, J.; Smith, J.S.; Byrnes, A.P. Clearance of adenovirus by Kupffer cells is mediated by scavenger receptors, natural antibodies, and complement. J. Virol. 2008, 82, 11705–11713. [Google Scholar]
- Parker, A.L.; Waddington, S.N.; Nicol, C.G.; Shayakhmetov, D.M.; Buckley, S.M.; Denby, L.; Kemball-Cook, G.; Ni, S.; Lieber, A.; McVey, J.H.; et al. Multiple vitamin K-dependent coagulation zymogens promote adenovirus-mediated gene delivery to hepatocytes. Blood 2006, 108, 2554–2561. [Google Scholar]
- Shayakhmetov, D.M.; Gaggar, A.; Ni, S.; Li, Z.Y.; Lieber, A. Adenovirus binding to blood factors results in liver cell infection and hepatotoxicity. J. Virol. 2005, 79, 7478–7491. [Google Scholar]
- Waddington, S.N.; McVey, J.H.; Bhella, D.; Parker, A.L.; Barker, K.; Atoda, H.; Pink, R.; Buckley, S.M.; Greig, J.A.; Denby, L.; et al. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell 2008, 132, 397–409. [Google Scholar]
- Parker, A.L.; Waddington, S.N.; Buckley, S.M.; Custers, J.; Havenga, M.J.; van Rooijen, N.; Goudsmit, J.; McVey, J.H.; Nicklin, S.A.; Baker, A.H. Effect of neutralizing sera on factor x-mediated adenovirus serotype 5 gene transfer. J. Virol. 2009, 83, 479–483. [Google Scholar]
- Kalyuzhniy, O.; Di Paolo, N.C.; Silvestry, M.; Hofherr, S.E.; Barry, M.A.; Stewart, P.L.; Shayakhmetov, D.M. Adenovirus serotype 5 hexon is critical for virus infection of hepatocytes in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 5483–5488. [Google Scholar]
- Prill, J.M.; Espenlaub, S.; Samen, U.; Engler, T.; Schmidt, E.; Vetrini, F.; Rosewell, A.; Grove, N.; Palmer, D.; Ng, P.; et al. Modifications of adenovirus hexon allow for either hepatocyte detargeting or targeting with potential evasion from Kupffer cells. Mol. Ther. 2011, 19, 83–92. [Google Scholar]
- Yoneyama, M.; Fujita, T. Recognition of viral nucleic acids in innate immunity. Rev. Med. Virol. 2010, 20, 4–22. [Google Scholar]
- Basner-Tschakarjan, E.; Gaffal, E.; O'Keeffe, M.; Tormo, D.; Limmer, A.; Wagner, H.; Hochrein, H.; Tuting, T. Adenovirus efficiently transduces plasmacytoid dendritic cells resulting in TLR9-dependent maturation and IFN-alpha production. J. Gene Med. 2006, 8, 1300–1306. [Google Scholar]
- Zhu, J.; Huang, X.; Yang, Y. Innate immune response to adenoviral vectors is mediated by both Toll-like receptor-dependent and -independent pathways. J. Virol. 2007, 81, 3170–3180. [Google Scholar]
- Yoneyama, M.; Kikuchi, M.; Natsukawa, T.; Shinobu, N.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Akira, S.; Fujita, T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004, 5, 730–737. [Google Scholar]
- Yoneyama, M.; Kikuchi, M.; Matsumoto, K.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Foy, E.; Loo, Y.M.; Gale, M., Jr.; Akira, S.; et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005, 175, 2851–2858. [Google Scholar]
- Andrejeva, J.; Childs, K.S.; Young, D.F.; Carlos, T.S.; Stock, N.; Goodbourn, S.; Randall, R.E. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 2004, 101, 17264–17269. [Google Scholar]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441, 101–105. [Google Scholar]
- Kato, H.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Uematsu, S.; Matsui, K.; Tsujimura, T.; Takeda, K.; Fujita, T.; Takeuchi, O.; et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity 2005, 23, 19–28. [Google Scholar]
- Hornung, V.; Ellegast, J.; Kim, S.; Brzozka, K.; Jung, A.; Kato, H.; Poeck, H.; Akira, S.; Conzelmann, K.K.; Schlee, M.; et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science 2006, 314, 994–997. [Google Scholar]
- Cui, S.; Eisenacher, K.; Kirchhofer, A.; Brzozka, K.; Lammens, A.; Lammens, K.; Fujita, T.; Conzelmann, K.K.; Krug, A.; Hopfner, K.P. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 2008, 29, 169–179. [Google Scholar]
- Nallagatla, S.R.; Hwang, J.; Toroney, R.; Zheng, X.; Cameron, C.E.; Bevilacqua, P.C. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science 2007, 318, 1455–1458. [Google Scholar]
- Kawai, T.; Takahashi, K.; Sato, S.; Coban, C.; Kumar, H.; Kato, H.; Ishii, K.J.; Takeuchi, O.; Akira, S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005, 6, 981–988. [Google Scholar]
- Mathews, M.B.; Shenk, T. Adenovirus virus-associated RNA and translation control. J. Virol. 1991, 65, 5657–5662. [Google Scholar]
- Fowlkes, D.M.; Shenk, T. Transcriptional control regions of the adenovirus VAI RNA gene. Cell 1980, 22, 405–413. [Google Scholar]
- Bhat, R.A.; Metz, B.; Thimmappaya, B. Organization of the noncontiguous promoter components of adenovirus VAI RNA gene is strikingly similar to that of eucaryotic tRNA genes. Mol. Cell. Biol. 1983, 3, 1996–2005. [Google Scholar]
- Akusjarvi, G.; Mathews, M.B.; Andersson, P.; Vennstrom, B.; Pettersson, U. Structure of genes for virus-associated RNAI and RNAII of adenovirus type 2. Proc. Natl. Acad. Sci. USA 1980, 77, 2424–2428. [Google Scholar]
- Ma, Y.; Mathews, M.B. Structure, function, and evolution of adenovirus-associated RNA: a phylogenetic approach. J. Virol. 1996, 70, 5083–5099. [Google Scholar]
- Clarke, P.A.; Pe'ery, T.; Ma, Y.; Mathews, M.B. Structural features of adenovirus 2 virus-associated RNA required for binding to the protein kinase DAI. Nucleic Acids Res. 1994, 22, 4364–4374. [Google Scholar]
- Ghadge, G.D.; Swaminathan, S.; Katze, M.G.; Thimmapaya, B. Binding of the adenovirus VAI RNA to the interferon-induced 68-kDa protein kinase correlates with function. Proc. Natl. Acad. Sci. USA 1991, 88, 7140–7144. [Google Scholar]
- Mellits, K.H.; Pe'ery, T.; Mathews, M.B. Role of the apical stem in maintaining the structure and function of adenovirus virus-associated RNA. J. Virol. 1992, 66, 2369–2377. [Google Scholar]
- Mellits, K.H.; Kostura, M.; Mathews, M.B. Interaction of adenovirus VA RNAl with the protein kinase DAI: nonequivalence of binding and function. Cell 1990, 61, 843–852. [Google Scholar]
- Mellits, K.H.; Mathews, M.B. Effects of mutations in stem and loop regions on the structure and function of adenovirus VA RNAI. Embo J. 1988, 7, 2849–2859. [Google Scholar]
- Ghadge, G.D.; Malhotra, P.; Furtado, M.R.; Dhar, R.; Thimmapaya, B. In vitro analysis of virus-associated RNA I (VAI RNA): inhibition of the double-stranded RNA-activated protein kinase PKR by VAI RNA mutants correlates with the in vivo phenotype and the structural integrity of the central domain. J. Virol. 1994, 68, 4137–4151. [Google Scholar]
- Pe'ery, T.; Mellits, K.H.; Mathews, M.B. Mutational analysis of the central domain of adenovirus virus-associated RNA mandates a revision of the proposed secondary structure. J. Virol. 1993, 67, 3534–3543. [Google Scholar]
- Rahman, A.; Malhotra, P.; Dhar, R.; Kewalramani, T.; Thimmapaya, B. Effect of single-base substitutions in the central domain of virus-associated RNA I on its function. J. Virol. 1995, 69, 4299–4307. [Google Scholar]
- Gwizdek, C.; Ossareh-Nazari, B.; Brownawell, A.M.; Doglio, A.; Bertrand, E.; Macara, I.G.; Dargemont, C. Exportin-5 mediates nuclear export of minihelix-containing RNAs. J. Biol. Chem. 2003, 278, 5505–5508. [Google Scholar]
- Subramanian, S.; Bhat, R.A.; Rundell, M.K.; Thimmappaya, B. Suppression of the translation defect phenotype specific for a virus-associated RNA-deficient adenovirus mutant in monkey cells by simian virus 40. J. Virol. 1986, 60, 363–368. [Google Scholar]
- Thimmappaya, B.; Weinberger, C.; Schneider, R.J.; Shenk, T. Adenovirus VAI RNA is required for efficient translation of viral mRNAs at late times after infection. Cell 1982, 31, 543–551. [Google Scholar]
- Aparicio, O.; Razquin, N.; Zaratiegui, M.; Narvaiza, I.; Fortes, P. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J. Virol. 2006, 80, 1376–1384. [Google Scholar]
- Bhat, R.A.; Thimmappaya, B. Adenovirus mutants with DNA sequence perturbations in the intragenic promoter of VAI RNA gene allow the enhanced transcription of VAII RNA gene in HeLa cells. Nucleic Acids Res. 1984, 12, 7377–7388. [Google Scholar]
- Kitajewski, J.; Schneider, R.J.; Safer, B.; Munemitsu, S.M.; Samuel, C.E.; Thimmappaya, B.; Shenk, T. Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2 alpha kinase. Cell 1986, 45, 195–200. [Google Scholar]
- Garcia, M.A.; Gil, J.; Ventoso, I.; Guerra, S.; Domingo, E.; Rivas, C.; Esteban, M. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 2006, 70, 1032–1060. [Google Scholar]
- Krishnamoorthy, T.; Pavitt, G.D.; Zhang, F.; Dever, T.E.; Hinnebusch, A.G. Tight binding of the phosphorylated alpha subunit of initiation factor 2 (eIF2alpha) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol. Cell. Biol. 2001, 21, 5018–5030. [Google Scholar]
- Nallagatla, S.R.; Toroney, R.; Bevilacqua, P.C. Regulation of innate immunity through RNA structure and the protein kinase PKR. Curr. Opin. Struct. Biol. 2011, 21, 119–127. [Google Scholar]
- Pathak, V.K.; Schindler, D.; Hershey, J.W. Generation of a mutant form of protein synthesis initiation factor eIF-2 lacking the site of phosphorylation by eIF-2 kinases. Mol. Cell. Biol. 1988, 8, 993–995. [Google Scholar]
- Rowlands, A.G.; Panniers, R.; Henshaw, E.C. The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J. Biol. Chem. 1988, 263, 5526–5533. [Google Scholar]
- O'Malley, R.P.; Mariano, T.M.; Siekierka, J.; Mathews, M.B. A mechanism for the control of protein synthesis by adenovirus VA RNAI. Cell 1986, 44, 391–400. [Google Scholar]
- Wahid, A.M.; Coventry, V.K.; Conn, G.L. Systematic deletion of the adenovirus-associated RNAI terminal stem reveals a surprisingly active RNA inhibitor of double-stranded RNA-activated protein kinase. J. Biol. Chem. 2008, 283, 17485–17493. [Google Scholar]
- Coventry, V.K.; Conn, G.L. Analysis of adenovirus VA RNAI structure and stability using compensatory base pair modifications. Nucleic Acids Res. 2008, 36, 1645–1653. [Google Scholar]
- Ma, Y.; Mathews, M.B. Comparative analysis of the structure and function of adenovirus virus-associated RNAs. J. Virol. 1993, 67, 6605–6617. [Google Scholar]
- Sano, M.; Kato, Y.; Taira, K. Sequence-specific interference by small RNAs derived from adenovirus VAI RNA. FEBS Lett. 2006, 580, 1553–1564. [Google Scholar]
- Lu, S.; Cullen, B.R. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J. Virol. 2004, 78, 12868–12876. [Google Scholar]
- Xu, N.; Segerman, B.; Zhou, X.; Akusjarvi, G. Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the rna-induced silencing complex and associate with polyribosomes. J. Virol. 2007, 81, 10540–10549. [Google Scholar]
- Andersson, M.G.; Haasnoot, P.C.; Xu, N.; Berenjian, S.; Berkhout, B.; Akusjarvi, G. Suppression of RNA interference by adenovirus virus-associated RNA. J. Virol. 2005, 79, 9556–9565. [Google Scholar]
- Aparicio, O.; Carnero, E.; Abad, X.; Razquin, N.; Guruceaga, E.; Segura, V.; Fortes, P. Adenovirus VA RNA-derived miRNAs target cellular genes involved in cell growth, gene expression and DNA repair. Nucleic Acids Res. 2010, 38, 750–763. [Google Scholar]
- Xu, N.; Gkountela, S.; Saeed, K.; Akusjarvi, G. The 5′-end heterogeneity of adenovirus virus-associated RNAI contributes to the asymmetric guide strand incorporation into the RNA-induced silencing complex. Nucleic Acids Res. 2009, 37, 6950–6959. [Google Scholar]
- Thimmappaya, B.; Jones, N.; Shenk, T. A mutation which alters initiation of transcription by RNA polymerase III on the Ad5 chromosome. Cell 1979, 18, 947–954. [Google Scholar]
- Vennstrom, B.; Pettersson, U.; Philipson, L. Two initiation sites for adenovirus 5.5S RNA. Nucleic Acids Res. 1978, 5, 195–204. [Google Scholar]
- Skalsky, R.L.; Cullen, B.R. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 2010, 64, 123–41. [Google Scholar]
- Cullen, B.R. Viral RNAs: lessons from the enemy. Cell 2009, 136, 592–597. [Google Scholar]
- Samanta, M.; Iwakiri, D.; Kanda, T.; Imaizumi, T.; Takada, K. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. Embo J. 2006, 25, 4207–4214. [Google Scholar]
- Iwakiri, D.; Zhou, L.; Samanta, M.; Matsumoto, M.; Ebihara, T.; Seya, T.; Imai, S.; Fujieda, M.; Kawa, K.; Takada, K. Epstein-Barr virus (EBV)-encoded small RNA is released from EBV-infected cells and activates signaling from Toll-like receptor 3. J. Exp. Med. 2009, 206, 2091–2099. [Google Scholar]
- Pichlmair, A.; Schulz, O.; Tan, C.P.; Naslund, T.I.; Liljestrom, P.; Weber, F.; Reis e Sousa, C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 2006, 314, 997–1001. [Google Scholar]
- Rajan, P.; Swaminathan, S.; Zhu, J.; Cole, C.N.; Barber, G.; Tevethia, M.J.; Thimmapaya, B. A novel translational regulation function for the simian virus 40 large-T antigen gene. J. Virol. 1995, 69, 785–795. [Google Scholar]
- Cascalló, M.; Capellà, G.; Mazo, A.; Alemany, R. Ras-dependent oncolysis with an adenovirus VAI mutant. Cancer Res. 2003, 63, 5544–5550. [Google Scholar]
- Machitani, M.; Katayama, K.; Sakurai, F.; Matsui, H.; Yamaguchi, T.; Suzuki, T.; Miyoshi, H.; Kawabata, K.; Mizuguchi, H. Development of an adenovirus vector lacking the expression of virus-associated RNAs. J. Control. Release 2011. In Press. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Machitani, M.; Yamaguchi, T.; Shimizu, K.; Sakurai, F.; Katayama, K.; Kawabata, K.; Mizuguchi, H. Adenovirus Vector-Derived VA-RNA-Mediated Innate Immune Responses. Pharmaceutics 2011, 3, 338-353. https://doi.org/10.3390/pharmaceutics3030338
Machitani M, Yamaguchi T, Shimizu K, Sakurai F, Katayama K, Kawabata K, Mizuguchi H. Adenovirus Vector-Derived VA-RNA-Mediated Innate Immune Responses. Pharmaceutics. 2011; 3(3):338-353. https://doi.org/10.3390/pharmaceutics3030338
Chicago/Turabian StyleMachitani, Mitsuhiro, Tomoko Yamaguchi, Kahori Shimizu, Fuminori Sakurai, Kazufumi Katayama, Kenji Kawabata, and Hiroyuki Mizuguchi. 2011. "Adenovirus Vector-Derived VA-RNA-Mediated Innate Immune Responses" Pharmaceutics 3, no. 3: 338-353. https://doi.org/10.3390/pharmaceutics3030338
APA StyleMachitani, M., Yamaguchi, T., Shimizu, K., Sakurai, F., Katayama, K., Kawabata, K., & Mizuguchi, H. (2011). Adenovirus Vector-Derived VA-RNA-Mediated Innate Immune Responses. Pharmaceutics, 3(3), 338-353. https://doi.org/10.3390/pharmaceutics3030338



