Novel Fri1-like Viruses Infecting Acinetobacter baumannii—vB_AbaP_AS11 and vB_AbaP_AS12—Characterization, Comparative Genomic Analysis, and Host-Recognition Strategy.
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Their Identification and Characterization
2.2. Phage Isolation, Propagation and Purification
2.3. Phage Host Range Determination
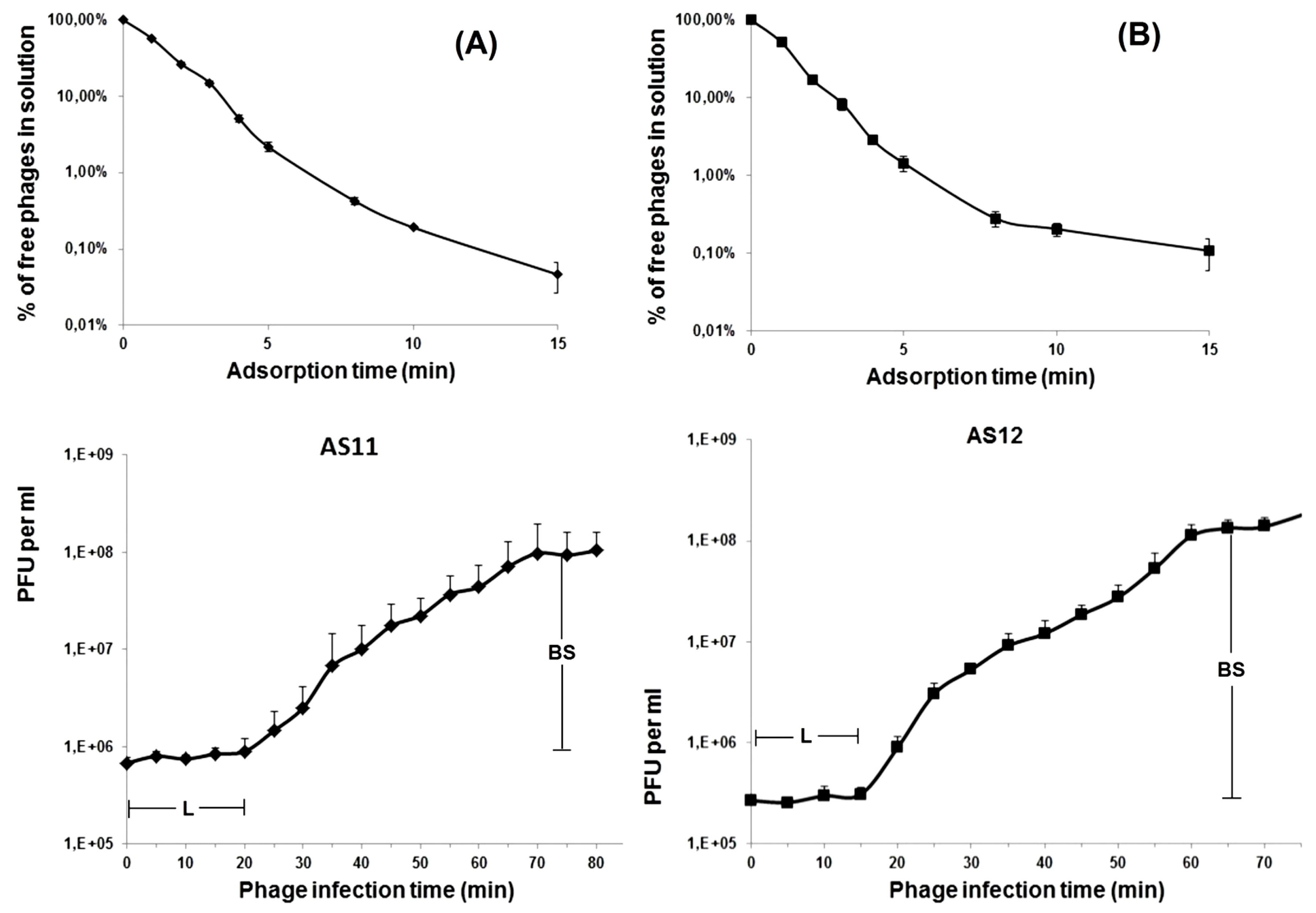
2.4. Phage Adsorption and One-Step Growth Experiments
2.5. Electron Microscopy
2.6. DNA Isolation and Sequencing
2.7. Genome Analysis
2.8. RNA Purification
2.9. Primer Extension and Manual DNA Sequencing
2.10. Nucleotide Sequence Accession Numbers
3. Results and Discussion
3.1. Phage Morphological Characteristics, Spectra of Lytic Activity and Infection Parameters
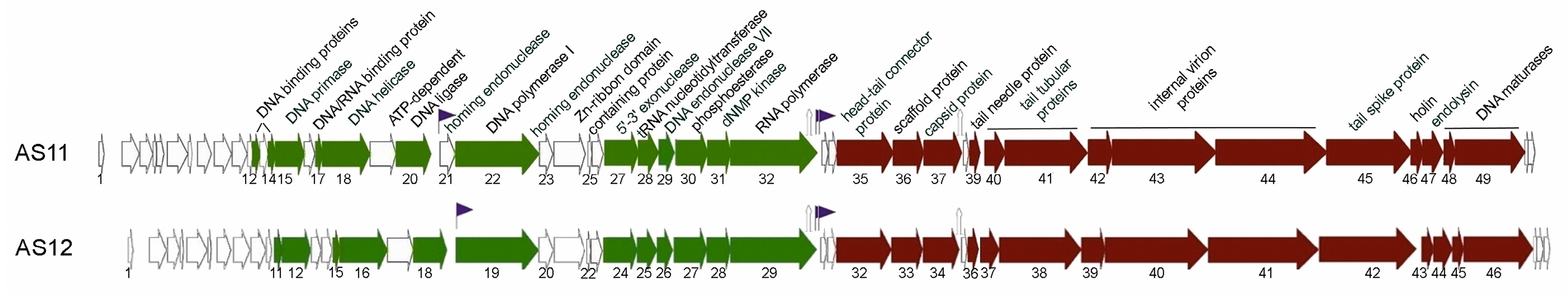
3.2. AS11 and AS12 Genome Organisation and Comparative Genomic Analysis
3.2.1. Early Genome Regions
3.2.2. Middle Genome Regions
3.2.3. Late Genome Regions
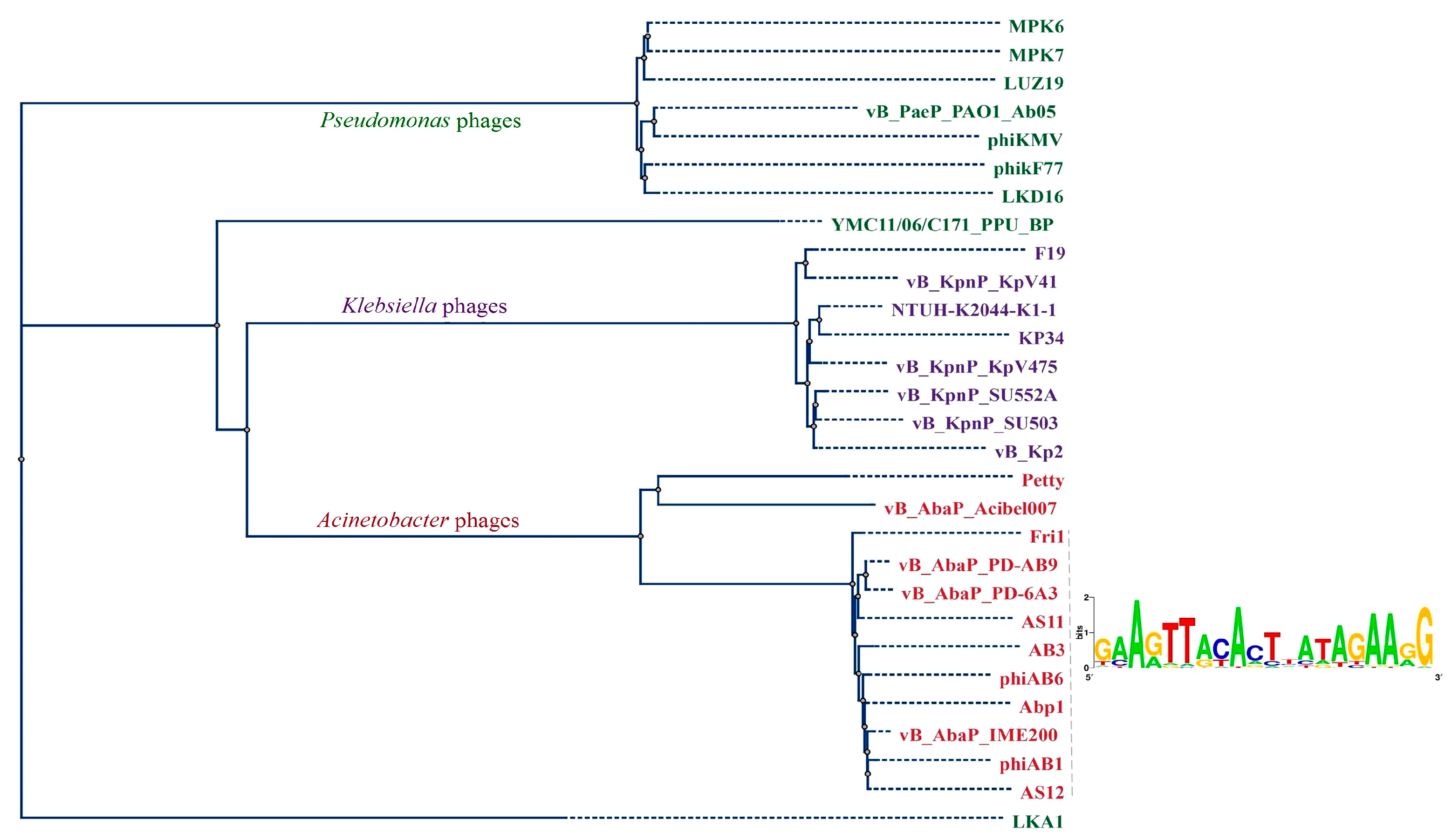
3.2.4. Comparative Genomics
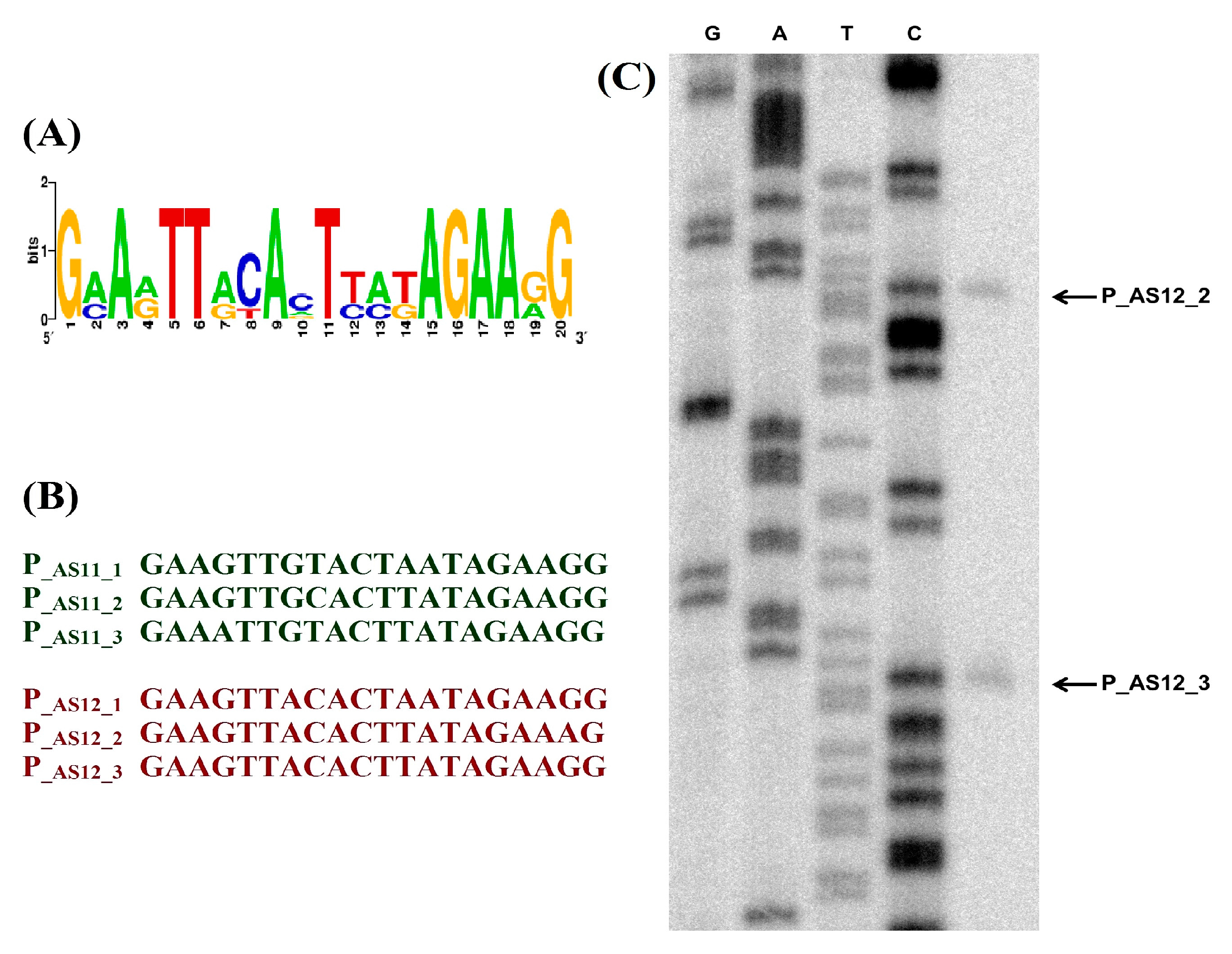
3.3. Regulation of Transcription
3.4. Phage Host Recognition Proteins and Primary A. baumannii Bacterial Receptors
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef] [PubMed]
- Towner, K.J. Acinetobacter: An old friend, but a new enemy. J. Hosp. Infect. 2009, 73, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Popova, A.V.; Zhilenkov, E.L.; Myakinina, V.P.; Krasilnikova, V.M.; Volozhantsev, N.V. Isolation and characterization of wide host range lytic bacteriophage AP22 infecting Acinetobacter baumannii. FEMS Microbiol. Lett. 2012, 332, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Li, Z.J.; Wang, S.W.; Wang, S.M.; Huang, D.H.; Li, Y.H.; Ma, Y.Y.; Wang, J.; Liu, F.; Chen, X.D.; et al. Isolation and characterization of ZZ1, a novel lytic phage that infects Acinetobacter baumannii clinical isolates. BMC Microbiol. 2012, 12, 156. [Google Scholar] [CrossRef] [PubMed]
- Merabishvili, M.; Vandenheuvel, D.; Kropinski, A.M.; Mast, J.; De Vos, D.; Verbeken, G.; Noben, J.P.; Lavigne, R.; Vaneechoutte, M.; Pirnay, J.P. Characterization of newly isolated lytic bacteriophages active against Acinetobacter baumannii. PLoS ONE 2014, 9, e104853. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.T.; Chiou, P.Y.; Chang, K.C.; Chen, L.K.; Lai, M.J. Isolation and characterization of phiAB2: A novel bacteriophage of Acinetobacter baumannii. Res. Microbiol. 2010, 161, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Le, S.; Peng, Y.; Zhao, Y.; Yin, S.; Zhang, L.; Yao, X.; Tan, Y.; Li, M.; Hu, F. Characterization and genome sequencing of phage Abp1, a new phiKMV-like virus infecting multidrug-resistant Acinetobacter baumannii. Curr. Microbiol. 2013, 66, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.J.; Chang, K.C.; Huang, S.W.; Luo, C.H.; Chiou, P.Y.; Wu, C.C.; Lin, N.T. The tail associated protein of Acinetobacter baumannii phage ΦAB6 is the host specificity determinant possessing exopolysaccharide depolymerase activity. PLoS ONE 2016, 11, e0153361. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.M.; Krupovic, M.; Knezevic, P.; Ackermann, H.W.; Barylski, J.; Brister, J.R.; Clokie, M.R.; Duffy, S.; Dutilh, B.E.; Edwards, R.A.; et al. Taxonomy of prokaryotic viruses: 2016 update from the ICTV bacterial and archaeal viruses subcommittee. Arch. Virol. 2017, 162, 1153–1157. [Google Scholar] [CrossRef] [PubMed]
- Sukhorukova, M.V.; Edelstein, M.V.; Skleenova, E.Y.; Ivanchik, N.V.; Mikotina, A.V.; Dekhnich, A.V.; Kozlov, R.S.; the «MARATHON» study group. Antimicrobial resistance of nosocomial Acinetobacter spp. isolates in Russia: Results of multicenter epidemiological study «MARATHON» 2013–2014. Clin. Microbiol. Antimicrob. Chemother. 2017, 19, 42–48. [Google Scholar]
- Bartual, S.G.; Seifert, H.; Hippler, C.; Luzon, M.A.; Wisplinghoff, H.; Rodríguez-Valera, F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 2005, 43, 4382–4390. [Google Scholar] [CrossRef] [PubMed]
- Diancourt, L.; Passet, V.; Nemec, A.; Dijkshoorn, L.; Brisse, S. The population structure of Acinetobacter baumannii: Expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS ONE 2010, 5, e10034. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.D. Bacteriophages; Interscience Publishers, Inc.: New York, NY, USA, 1959; OCLC 326505. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular cloning: A laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989; ISBN 0-87969-309-6. [Google Scholar]
- Brenner, S.; Horne, R.W. A negative staining method for high resolution electron microscopy of viruses. Biochim. Biophys. Acta 1959, 34, 103–110. [Google Scholar] [CrossRef]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. Methods Mol. Biol. 2000, 132, 71–91. [Google Scholar] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genomics 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; et al. CDD: Specific functional annotation with the Conserved DomainDatabase. Nucleic Acids Res. 2009, 37, D205–D210. [Google Scholar] [CrossRef] [PubMed]
- Söding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–248. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Arnold Finding Terminators. Available online: http://rna.igmors.u-psud.fr/toolbox/arnold/ (accessed on 17 November 2016).
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Clustal Omega. Available online: http://www.ebi.ac.uk/Tools/msa/clustalo/ (accessed on 21 April 2017).
- Molecular evolution, phylogenetics and epidemiology. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 21 April 2017).
- Alikhan, N-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Dubrovin, E.V.; Popova, A.V.; Kraevskiy, S.V.; Ignatov, S.G.; Ignatyuk, T.E.; Yaminsky, I.V.; Volozhantsev, N.V. Atomic force microscopy analysis of the Acinetobacter baumannii bacteriophage AP22 lytic cycle. PLoS ONE 2012, 7, e47348. [Google Scholar] [CrossRef] [PubMed]
- Kitti, T.; Thummeepak, R.; Thanwisai, A.; Boonyodying, K.; Kunthalert, D.; Ritvirool, P.; Sitthisak, S. Characterization and detection of endolysin gene from three Acinetobacter baumannii bacteriophages isolated from sewage water. Indian J. Microbiol. 2014, 54, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Cornelissen, A.; Ceyssens, P-J.; Krylov, V.N.; Noben, J-P.; Volckaert, G.; Lavigne, R. Identification of EPS-degrading activity within the tail spikes of the novel Pseudomonas putida phage AF. Virology 2012, 434, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Harper, D.; Parracho, H.; Walker, J.; Sharp, R.; Hughes, G.; Werthén, M.; Lehman, S.; Morales, S. Bacteriophages and biofilms. Antibiotics 2014, 3, 270–284. [Google Scholar] [CrossRef]
- Hsu, C-R.; Lin, T-L.; Pan, Y-J.; Hsieh, P-F.; Wang, J-T. Isolation of a bacteriophage specific for a new capsular type of Klebsiella pneumoniae and characterization of its polysaccharide depolymerase. PLoS ONE 2013, 8, e70092. [Google Scholar] [CrossRef] [PubMed]
- Hughes, K.A.; Sutherland, I.W.; Clark, J.; Jones, M.V. Bacteriophage and associated polysaccharide depolymerases-novel tools for study of bacterial biofilms. J Appl Microbiol. 1998, 85, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Oliveira, H.; Melo, L.D.; Sillankorva, S.; Azeredo, J. Bacteriophage-encoded depolymerases: Their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef] [PubMed]
- Farrugia, D.N.; Elbourne, L.D.H.; Hassan, K.A.; Eijkelkamp, B.A.; Tetu, S.G.; et al. The Complete Genome and Phenome of a Community-Acquired Acinetobacter baumannii. PLoS ONE 2013, 8, e58628. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, R.; Lecoutere, E.; Wagemans, J.; Cenens, W.; Aertsen, A.; Schoofs, L.; Landuyt, B.; Paeshuyse, J.; Scheer, M.; Schobert, M.; Ceyssens, P.J. A multifaceted study of Pseudomonas aeruginosa shutdown by virulent podovirus LUZ19. MBio. 2013, 4, e00061-13. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, R.; Burkal’tseva, M.V.; Robben, J.; Sykilinda, N.N.; Kurochkina, L.P.; Grymonprez, B.; Jonckx, B.; Krylov, V.N.; Mesyanzhinov, V.V.; Volckaert, G. The genome of bacteriophage phiKMV, a T7-like virus infecting Pseudomonas aeruginosa. Virology 2003, 312, 49–59. [Google Scholar] [CrossRef]
- Ceyssens, P-J.; Lavigne, R.; Mattheus, W.; Chibeu, A.; Hertveldt, K.; Mast, J.; Robben, J.; Volckaert, G. Genomic analysis of Pseudomonas aeruginosa phages LKD16 and LKA1: Establishment of the phiKMV subgroup within the T7 supergroup. J. Bacteriol. 2006, 188, 6924–6931. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.C.; Lin, N.T.; Hu, A.; Lin, Y.S.; Chen, L.K.; Lai, M.J. Genomic analysis of bacteriophage ϕAB1, a ϕKMV-like virus infecting multidrug-resistant Acinetobacter baumannii. Genomics 2011, 97, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Young, R.; Wang, I.-N.; Roof, W.D. Phages will out: Strategies of host cell lysis. Trends Microbiol. 2000, 8, 120–128. [Google Scholar] [CrossRef]
- Hesselbach, B.A.; Nakada, D. Host shutoff function of bacteriophage T7: Involvement of T7 gene 2 and gene 0.7 in the inactivation of Escherichia coli RNA polymerase. J. Virol. 1977, 24, 736–745. [Google Scholar] [PubMed]
- Nechaev, S.; Severinov, K. Inhibition of Escherichia coli RNA polymerase by bacteriophage T7 gene 2 protein. J. Mol. Biol. 1999, 289, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Klimuk, E.; Akulenko, N.; Makarova, KS.; Ceyssens, P-J.; Lavigne, R.; Severinov, K. Host RNA polymerase inhibitors encoded by φKMV-like phages of Pseudomonas. Virology 2013, 436, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Savalia, D.; Nechaev, S.; Robins, R.; Molineux, I.; Severinov, K. The role of the T7 Gp2 inhibitor of host RNA polymerase in phage development. J. Mol. Biol. 2010, 402, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Kemp, P.; Gupta, M.; Molineux, I.J. Bacteriophage T7 DNA ejection into cells is initiated by an enzyme-like mechanism. Mol. Microbiol. 2004, 53, 1251–1265. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.M.; Tu, I.F.; Yang, F.L.; Ko, T.P.; Liao, J.H.; Lin, N.T.; Wu, C.Y.; Ren, C.T.; Wang, A.H.; Chang, C.M.; et al. Structural basis for fragmenting the exopolysaccharide of Acinetobacter baumannii by bacteriophage phiAB6 tailspike protein. Sci. Rep. 2017, 7, 42711. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, J.J.; Shneider, M.M.; Senchenkova, S.N.; Shashkov, A.S.; Siniagina, M.N.; Malanin, S.Y.; Popova, A.V.; Miroshnikov, K.A.; Hall, R.M.; Knirel, Y.A. The K19 capsular polysaccharide of Acinetobacter baumannii is produced via a Wzy polymerase encoded in a small genomic island rather than the KL19 capsule gene cluster. Microbiology 2016, 162, 1479–1489. [Google Scholar] [CrossRef] [PubMed]
- Shashkov, A.S.; Kenyon, J.J.; Senchenkova, S.N.; Shneider, M.M.; Popova, A.V.; Arbatsky, N.P.; Miroshnikov, K.A.; Volozhantsev, N.V.; Hall, R.M.; Knirel, Y.A. Acinetobacter baumannii K27 and K44 capsular polysaccharides have the same K unit but different structures due to the presence of distinct wzy genes in otherwise closely related K gene clusters. Glycobiology 2016, 26, 501–508. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Popova, A.V.; Lavysh, D.G.; Klimuk, E.I.; Edelstein, M.V.; Bogun, A.G.; Shneider, M.M.; Goncharov, A.E.; Leonov, S.V.; Severinov, K.V. Novel Fri1-like Viruses Infecting Acinetobacter baumannii—vB_AbaP_AS11 and vB_AbaP_AS12—Characterization, Comparative Genomic Analysis, and Host-Recognition Strategy. Viruses 2017, 9, 188. https://doi.org/10.3390/v9070188
Popova AV, Lavysh DG, Klimuk EI, Edelstein MV, Bogun AG, Shneider MM, Goncharov AE, Leonov SV, Severinov KV. Novel Fri1-like Viruses Infecting Acinetobacter baumannii—vB_AbaP_AS11 and vB_AbaP_AS12—Characterization, Comparative Genomic Analysis, and Host-Recognition Strategy. Viruses. 2017; 9(7):188. https://doi.org/10.3390/v9070188
Chicago/Turabian StylePopova, Anastasia V., Daria G. Lavysh, Evgeniy I. Klimuk, Mikhail V. Edelstein, Alexander G. Bogun, Mikhail M. Shneider, Artemiy E. Goncharov, Sergey V. Leonov, and Konstantin V. Severinov. 2017. "Novel Fri1-like Viruses Infecting Acinetobacter baumannii—vB_AbaP_AS11 and vB_AbaP_AS12—Characterization, Comparative Genomic Analysis, and Host-Recognition Strategy." Viruses 9, no. 7: 188. https://doi.org/10.3390/v9070188
APA StylePopova, A. V., Lavysh, D. G., Klimuk, E. I., Edelstein, M. V., Bogun, A. G., Shneider, M. M., Goncharov, A. E., Leonov, S. V., & Severinov, K. V. (2017). Novel Fri1-like Viruses Infecting Acinetobacter baumannii—vB_AbaP_AS11 and vB_AbaP_AS12—Characterization, Comparative Genomic Analysis, and Host-Recognition Strategy. Viruses, 9(7), 188. https://doi.org/10.3390/v9070188




