Variability of Emaravirus Species Associated with Sterility Mosaic Disease of Pigeonpea in India Provides Evidence of Segment Reassortment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of SMD-Affected Pigeonpea Leaf Samples
2.2. RNA Extraction, RT-PCR and Cloning of PPSMV-1 and PPSMV-2 Segments
2.3. Sequence Analysis of RNA Fragments of PPSMV-1 and PPSMV-2 Isolates
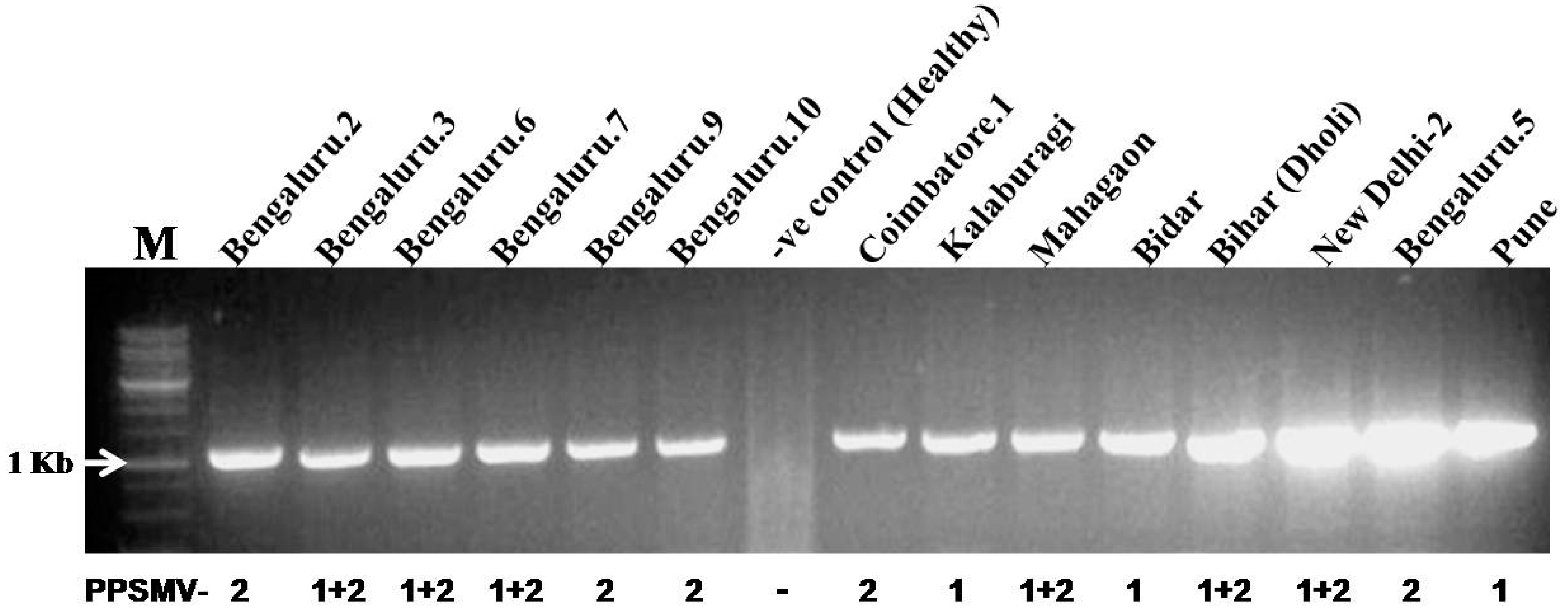
2.4. Diagnostic Multiplex-RT-PCR for Detection of PPSMV-1 and PPSMV-2 Isolates
3. Results
3.1. Analysis of NP and MP Sequence Identity between PPSMV-1 and PPSMV-2 Isolates
3.2. RNA6 is Also Associated with Isolates of PPSMV-1
3.3. Recombination Analysis for Sequences of PPSMV-1 and PPSMV-2 Isolates
3.4. Phylogenetic Analysis of PPSMV-1 and PPSMV-2 Sequences
3.5. Diagnostic Multiplex-RT-PCR and Distribution of PPSMV-1 and PPSMV-2
3.6. Reassortment of RNA4 Segment from PPSMV-1 to PPSMV-2 Isolates
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Elbeaino, T.; Digiaro, M.; Uppala, M.; Sudini, H. Deep sequencing of pigeonpea sterility mosaic virus discloses five RNA segments related to emaraviruses. Virus Res. 2014, 188, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Elbeaino, T.; Digiaro, M.; Uppala, M.; Sudini, H. Deep sequencing of dsRNAs recovered from mosaic-diseased pigeonpea reveals the presence of a novel emaravirus: Pigeonpea sterility mosaic virus 2. Arch. Virol. 2015, 160, 2019–2029. [Google Scholar] [CrossRef] [PubMed]
- Patil, B.L.; Kumar, P.L. Pigeonpea sterility mosaic virus: A legume-infecting Emaravirus from South Asia. Mol. Plant Pathol. 2015, 16, 775–786. [Google Scholar] [CrossRef] [PubMed]
- Mitra, M. Report of the Imperial Mycologist; Scientific Reports of the Agricultural Research Institute, Pusa; Superintendent of Government Printing: Calcutta, India, 1931; Volume 19, pp. 58–71. [Google Scholar]
- Reddy, M.V.; Raju, T.N.; Lenne, J.M. Diseases of pigeonpea. In The Pathology of Food and Pasture Legumes; Allen, D.J., Lenne, J.M., Eds.; CAB International: Wallingford, UK, 1998; pp. 517–558. [Google Scholar]
- Jones, A.T.; Kumar, P.L.; Saxena, K.B.; Kulkarni, N.K.; Muniyappa, V.; Waliyar, F. Sterility mosaic disease-the “green plague” of pigeonpea: Advances in understanding the etiology, transmission and control of a major virus disease. Plant Dis. 2004, 88, 436–445. [Google Scholar] [CrossRef]
- Kumar, P.L.; Jones, A.T.; Reddy, D.V.R. Mechanical transmission of Pigeonpea sterility mosaic virus. J. Mycol. Plant Pathol. 2002, 32, 88–89. [Google Scholar]
- Kumar, P.L.; Jones, A.T.; Reddy, D.V. A novel mite-transmitted virus with a divided RNA genome closely associated with pigeonpea sterility mosaic disease. Phytopathology 2003, 93, 71–81. [Google Scholar] [CrossRef] [PubMed]
- ICTV. 2016. Available online: https://talk.ictvonline.org/files/master-species-lists/m/msl/6776 (accessed on 5 July 2017).
- Mielke-Ehret, N.; Muehlbach, H.P. A novel, multipartite, negative-strand RNA virus is associated with the ringspot disease of European mountain ash (Sorbus aucuparia L.). J. Gen. Virol. 2007, 88, 1337–1346. [Google Scholar] [CrossRef] [PubMed]
- Mielke-Ehret, N.; Muhlbach, H.P. Emaravirus: A novel genus of multipartite, negative strand RNA plant viruses. Viruses 2012, 4, 1515–1536. [Google Scholar] [CrossRef] [PubMed]
- Di Bello, P.L.; Ho, T.; Tzanetakis, I.E. The evolution of emaraviruses is becoming more complex: Seven segments identified in the causal agent of Rose rosette disease. Virus Res. 2015, 210, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Di Bello, P.L.; Laney, A.G.; Druciarek, T.; Ho, T.; Gergerich, R.C.; Keller, K.E.; Martin, R.R.; Tzanetakis, I.E. A novel emaravirus is associated with redbud yellow ringspot disease. Virus Res. 2016, 222, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Laney, A.G.; Keller, K.E.; Martin, R.R.; Tzanetakis, I.E. A discovery 70 years in the making: Characterization of the Rose rosette virus. J. Gen. Virol. 2011, 92, 1727–1732. [Google Scholar] [CrossRef] [PubMed]
- Mühlbach, H.P.; Mielke-Ehret, N. Emaravirus. In Virus Taxonomy. Classification and Nomenclature of Viruses; Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkovitz, E.J., Eds.; Elsevier: Oxford, UK, 2011; pp. 767–770. [Google Scholar]
- Tatineni, S.; McMechan, A.J.; Wosula, E.N.; Wegulo, S.N.; Graybosch, R.A.; French, R.; Hein, G.L. An eriophyid mite-transmitted plant virus contains eight genomic RNA segments with unusual heterogeneity in the nucleocapsid protein. J. Virol. 2014, 88, 11834–11845. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Navarro, B.; Wang, G.; Wang, Y.; Yang, Z.; Xu, W.; Zhu, C.; Wang, L.; Serio, F.D.; Hong, N. Actinidia chlorotic ringspot-associated virus: A novel emaravirus infecting kiwifruit plants. Mol. Plant Pathol. 2016, 18. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Leal, R.; Duffy, S.; Xiong, Z.; Hammond, R.W.; Elena, S.F. Advances in plant virus evolution: Translating evolutionary insights into better disease management. Phytopathology 2011, 101, 1136–1148. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E.; Holland, J.J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997, 51, 151–178. [Google Scholar] [CrossRef] [PubMed]
- Kormelink, R.; Garcia, M.L.; Goodin, M.; Sasaya, T.; Haenni, A.L. Negative-strand RNA viruses: The plant-infecting counterparts. VirusRes. 2011, 162, 184–202. [Google Scholar] [CrossRef] [PubMed]
- McDonald, S.M.; Nelson, M.I.; Turner, P.E.; Patton, J.T. Reassortment in segmented RNA viruses: Mechanisms and outcomes. Nat. Rev. Microbiol. 2016, 14, 448–460. [Google Scholar] [CrossRef] [PubMed]
- Nagy, P.D. Recombination in plant RNA viruses. In Plant Virus Evolution; Roosinck, M.J., Ed.; Springer: Berlin, Germany, 2008; pp. 133–164. [Google Scholar]
- Drummond, A.J.; Pybus, O.G.; Rambaut, A.; Forsberg, R.; Rodrigo, A.G. Measurably evolving populations. Trends Ecol. Evolut. 2003, 18, 481–488. [Google Scholar] [CrossRef]
- García-Arenal, F.; Fraile, A.; Malpica, J.M. Variability and genetic structure of plant virus populations. Annu. Rev. Phytopathol. 2001, 39, 157–186. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, A.J.; Fargette, D.; Garcia-Arenal, F.; Gibbs, M.J. Time-The emerging dimension of plant virus studies. J. Gen. Virol. 2010, 91, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Lemmetty, A.; Latvala, S.; Samuilova, O.; Valkonen, J.P.T. Occurrence and genetic diversity of Raspberry leaf blotch virus (RLBV) infecting cultivated and wild Rubus species in Finland. Ann. Appl. Biol. 2016, 168, 122–132. [Google Scholar] [CrossRef]
- Kallinen, A.K.; Lindberg, I.L.; Tugume, A.K.; Valkonen, J.P. Detection, distribution, and genetic variability of European mountain ash ringspot-associated virus. Phytopathology 2009, 99, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Mbanzibwa, D.R.; Tian, Y.P.; Tugume, A.K.; Patil, B.L.; Yadav, J.S.; Bagewadi, B.; Abarshi, M.M.; Alicai, T.; Changadeya, W.; Mkumbira, J.; et al. Evolution of cassava brown streak disease-associated viruses. J. Gen. Virol. 2011, 92, 974–987. [Google Scholar] [CrossRef] [PubMed]
- Stewart, L.R. Sequence diversity of wheat mosaic virus isolates. Virus Res. 2016, 213, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Walia, J.J.; Salem, N.M.; Falk, B.W. Partial sequence and survey analysis identify a multipartite, negative-sense RNA virus associated with fig mosaic. Plant Dis. 2009, 93, 4–10. [Google Scholar] [CrossRef]
- Walia, J.J.; Willemsen, A.; Elci, E.; Caglayan, K.; Falk, B.W.; Rubio, L. Genetic variation and possible mechanisms driving the evolution of worldwide fig mosaic virus isolates. Phytopathology 2014, 104, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Elbeaino, T.; Whitfield, A.; Sharma, M.; Digiaro, M. Emaravirus-specific degenerate PCR primers allowed the identification of partial RNA-dependent RNA polymerase sequences of Maize red stripe virus and Pigeonpea sterility mosaic virus. J. Virol. Methods 2013, 188, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1. [Google Scholar] [CrossRef] [PubMed]
- Babu, B.; Washburn, B.K.; Poduch, K.; Knox, G.W.; Paret, M.L. Identification and characterization of two novel genomic RNA segments RNA5 and RNA6 in rose rosette virus infecting roses. Acta Virol. 2016, 60, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Elbeaino, T.; Digiaro, M.; Martelli, G.P. RNA-5 and -6, two additional negative-sense RNA segments associated with Fig mosaic virus. J. Plant Pathol. 2012, 94, 421–425. [Google Scholar]
- Lu, Y.; McGavin, W.; Cock, P.J.; Schnettler, E.; Yan, F.; Chen, J.; MacFarlane, S. Newly identified RNAs of raspberry leaf blotch virus encoding a related group of proteins. J. Gen. Virol. 2015, 96, 3432–3439. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.; Maejima, K.; Komatsu, K.; Kitazawa, Y.; Hashimoto, M.; Takata, D.; Yamaji, Y.; Namba, S. Identification and characterization of two novel genomic RNA segments of fig mosaic virus, RNA5 and RNA6. J. Gen. Virol. 2012, 93, 1612–1619. [Google Scholar] [CrossRef] [PubMed]
- Patil, B.L.; Ogwok, E.; Wagaba, H.; Mohammed, I.U.; Yadav, J.S.; Bagewadi, B.; Taylor, N.J.; Kreuze, J.F.; Maruthi, M.N.; Alicai, T.; et al. RNAi-mediated resistance to diverse isolates belonging to two virus species involved in Cassava brown streak disease. Mol. Plant Pathol. 2011, 12, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Tentchev, D.; Verdin, E.; Marchal, C.; Jacquet, M.; Aguilar, J.M.; Moury, B. Evolution and structure of Tomato spotted wilt virus populations: Evidence of extensive reassortment and insights into emergence processes. J. Gen. Virol. 2011, 92, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Briese, T.; Calisher, C.H.; Higgs, S. Viruses of the family Bunyaviridae: Are all available isolates reassortants? Virology 2013, 446, 207–216. [Google Scholar] [CrossRef] [PubMed]


| PPSMV Genomic Segment | RNA 1 | RNA 2 | RNA 4 | RNA 5 | ||
|---|---|---|---|---|---|---|
| Recombinant Isolate | PPSMV-1 Bihar | PPSMV-1 Pune | PPSMV-1 Mahagaon | PPSMV-2 Patancheru | PPSMV-1 Bihar | |
| Recombination Breakpoint | 5403–5495 nt | 554–1550 nt | 862–1178 nt | 1228–1398 nt | 453–688 nt | |
| Parent Isolates | Major | PPSMV-1 Mahagaon | PPSMV-1 Patancheru | PPSMV-1 Kalaburagi | PPSMV-1 Kalaburagi | PPSMV-2 Bihar |
| Minor | PPSMV-2 Patancheru | PPSMV-1 Kalaburagi | PPSMV-1 Delhi.1 | PPSMV-1 Patancheru | PPSMV-1 Kalaburagi | |
| p-values for 7 recombination detection methods of RDP4 | RDP | 8.710 × 10−07 | NS | 4.794 × 10−08 | 1.432 × 10−10 | NS |
| GENECONV | 3.152 × 10−07 | 1.811 × 10−02 | NS | 1.820 × 10−09 | 1.877 × 10−05 | |
| BOOTSCAN | NS | 6.993 × 10−03 | 4.917 × 10−02 | 1.134 × 10−10 | 1.748 × 10−06 | |
| MAXI CHISQUARE | NS | 6.874 × 10−04 | 2.057 ×10−02 | 1.043 ×10−03 | 1.556 × 10−04 | |
| CHIMAERA | NS | 2.252 × 10−04 | NS | 9.874×10−04 | 6.743 × 10−03 | |
| SISCAN | 2.912 × 10−08 | 2.404 × 10−06 | 3.634× 10−06 | 3.692 ×10−06 | 1.886 × 10−10 | |
| 3SEQ | 1.454 × 10−09 | 5.609 × 10−06 | 3.969 × 10−02 | NS | 2.209 × 10−04 | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patil, B.L.; Dangwal, M.; Mishra, R. Variability of Emaravirus Species Associated with Sterility Mosaic Disease of Pigeonpea in India Provides Evidence of Segment Reassortment. Viruses 2017, 9, 183. https://doi.org/10.3390/v9070183
Patil BL, Dangwal M, Mishra R. Variability of Emaravirus Species Associated with Sterility Mosaic Disease of Pigeonpea in India Provides Evidence of Segment Reassortment. Viruses. 2017; 9(7):183. https://doi.org/10.3390/v9070183
Chicago/Turabian StylePatil, Basavaprabhu L., Meenakshi Dangwal, and Ritesh Mishra. 2017. "Variability of Emaravirus Species Associated with Sterility Mosaic Disease of Pigeonpea in India Provides Evidence of Segment Reassortment" Viruses 9, no. 7: 183. https://doi.org/10.3390/v9070183





