Chloroviruses Have a Sweet Tooth
Abstract
:1. Introduction
2. Chlorovirus Encoded Polysaccharide Synthesizing Enzymes
3. Chlorovirus Encoded Nucleotide Sugar Metabolism Enzymes
4. Unusual Attachment of Glycans to the Chlorovirus Major Capsid Proteins
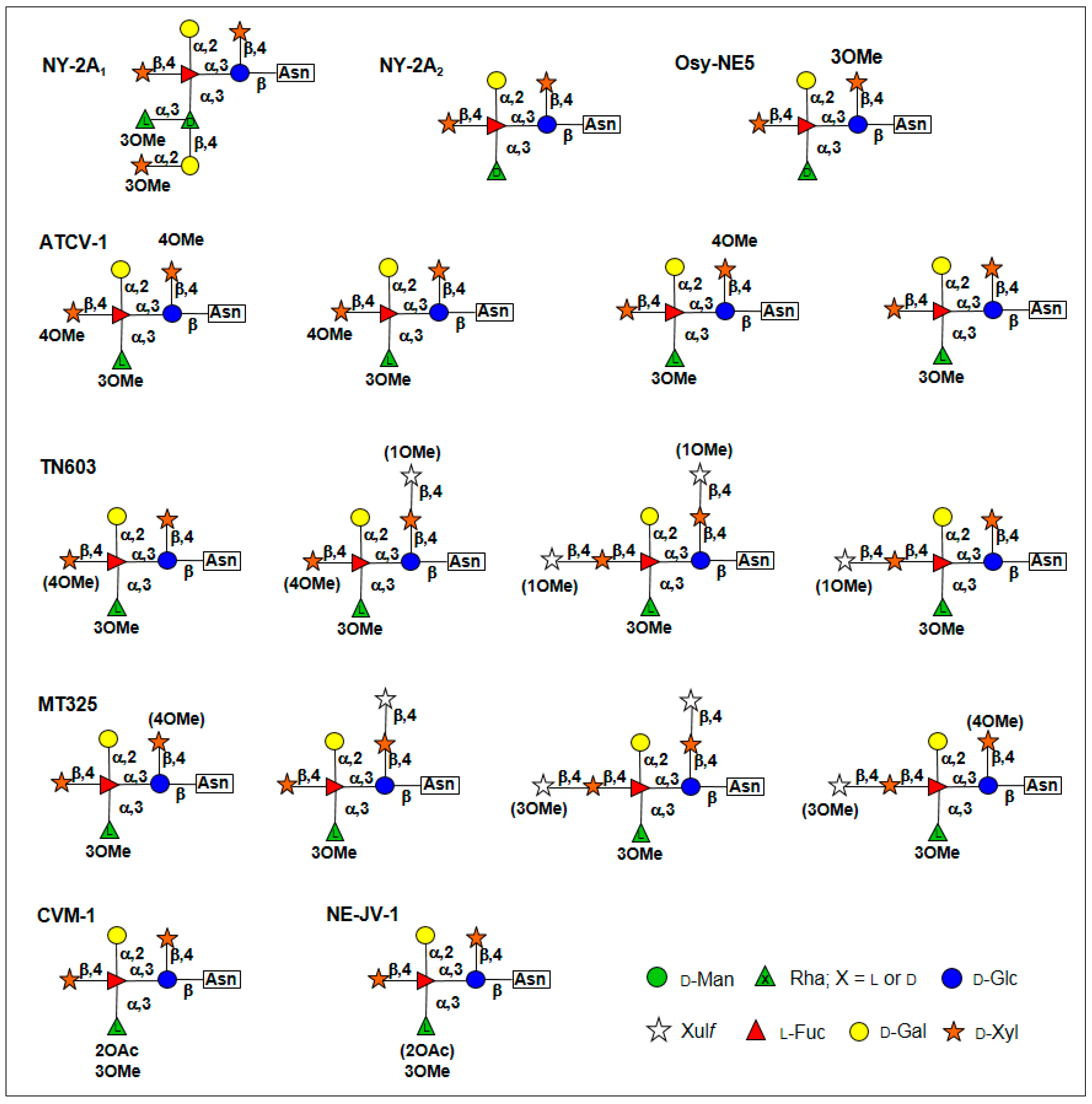
5. Glycan Structures Attached to Chlorovirus Major Capsid Proteins
6. Chlorovirus PBCV-1 Encoded Glycosyltransferases
7. Additional Chlorovirus Encoded Sugar Metabolism Enzymes
8. Chlorovirus-Encoded Polysaccharide Degrading Enzymes
9. Conservation of the Chlorovirus Encoded Sugar Enzymes
10. Sugar Enzymes Coded by Other Large DNA Viruses
Acknowledgments
Conflicts of Interest
References
- Karakashian, S.J.; Karakashian, M.W. Evolution and symbiosis in the genus Chlorella and related algae. Evolution 1965, 19, 368–377. [Google Scholar] [CrossRef]
- Kodama, Y.; Suzuki, H.; Dohra, H.; Sugii, M.; Kitazume, T.; Yamaguchi, K.; Shigenobu, S.; Fujishima, M. Comparison of gene expression of Paramecium bursaria with and without Chlorella variabilis symbionts. BMC Genomics 2014, 15, 1. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Onimatsu, H.; Van Etten, J.L. Chlorella viruses. In Advances in Virus Research; Maramorosch, K., Shatkin, A.J., Eds.; Elsevier Inc: San Diego, CA, USA, 2006; Vol. 66, pp. 293–366. [Google Scholar]
- Dunigan, D.D.; Fitzgerald, L.A.; Van Etten, J.L. Phycodnaviruses: a peek at genetic diversity. Virus Res. 2006, 117, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Van Etten, J.L.; Dunigan, D.D. Chloroviruses: not your everyday plant virus. Trends Plant Sci. 2012, 17, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Quispe, C.F.; Esmael, A.; Sonderman, O.; McQuinn, M.; Agarkova, I.; Battah, M.; Duncan, G.A.; Dunigan, D.D.; Smith, T.P.; De Castro, C.; et al. Characterization of a new chlorovirus type with permissive and non-permissive features on phylogenetically related algal strains. Virology 2017, 500, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, L.A.; Graves, M.V.; Li, X.; Feldblyum, T.; Hartigan, J.; Van Etten, J.L. Sequence and annotation of the 314-kb MT325 and the 321-kb FR483 viruses that infect Chlorella Pbi. Virology 2007, 358, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, L.A.; Graves, M.V.; Li, X.; Feldblyum, T.; Nierman, W.C.; Van Etten, J.L. Sequence and annotation of the 369-kb NY-2A and the 345-kb AR158 viruses that infect Chlorella NC64A. Virology 2007, 358, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, L.A.; Graves, M.V.; Li, X.; Hartigan, J.; Pfitzner, A.J.; Hoffart, E.; Van Etten, J.L. Sequence and annotation of the 288-kb ATCV-1 virus that infects an endosymbiotic chlorella strain of the heliozoon Acanthocystis turfacea. Virology 2007, 362, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Dunigan, D.D.; Cerny, R.L.; Bauman, A.T.; Roach, J.C.; Lane, L.C.; Agarkova, I.V.; Wulser, K.; Yanai-Balser, G.M.; Gurnon, J.R.; Vitek, J.C.; et al. Paramecium bursaria chlorella virus 1 proteome reveals novel architectural and regulatory features of a giant virus. J. Virol. 2012, 86, 8821–8834. [Google Scholar] [CrossRef] [PubMed]
- Jeanniard, A.; Dunigan, D.D.; Gurnon, J.R.; Agarkova, I.V.; Kang, M.; Vitek, J.; Duncan, G.; McClung, O.W.; Larsen, M.; Claverie, J.-M.; et al. Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses. BMC Genomics 2013, 14, 158. [Google Scholar] [CrossRef] [PubMed]
- Van Etten, J.L.; Burbank, D.E.; Kuczmarski, D.; Meints, R.H. Virus infection of culturable chlorella-like algae and development of a plaque assay. Science 1983, 219, 994–996. [Google Scholar] [CrossRef] [PubMed]
- Cherrier, M.V.; Kostyuchenko, V.A.; Xiao, C.; Bowman, V.D.; Battisti, A.J.; Yan, X.; Chipman, P.R.; Baker, T.S.; Van Etten, J.L.; Rossmann, M.G. An icosahedral algal virus has a complex unique vertex decorated by a spike. Proc. Natl. Acad. Sci. USA 2009, 106, 11085–11089. [Google Scholar] [CrossRef] [PubMed]
- Strasser, P.; Zhang, Y.P.; Rohozinski, J.; Van Etten, J.L. The termini of the chlorella virus PBCV-1 genome are identical 2.2-kbp inverted repeats. Virology 1991, 180, 763–769. [Google Scholar] [CrossRef]
- Zhang, Y.; Strasser, P.; Grabherr, R.; Van Etten, J.L. Hairpin loop structure at the termini of the chlorella virus PBCV-1 genome. Virology 1994, 202, 1079–1082. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.; Zhang, Y.; Van Etten, J.L. DNA methyltransferases and DNA site-specific endonucleases encoded by chlorella viruses. In DNA methylation: molecular biology and biological significance; Jost, J.P., Saluz, H.P., Eds.; Birkhauser Verlag Publ: Basel, Switzerland, 1993; pp. 186–211. [Google Scholar]
- DeAngelis, P.L.; Jing, W.; Graves, M.V.; Burbank, D.E.; Van Etten, J.L. Hyaluronan synthase of chlorella virus PBCV-1. Science 1997, 278, 1800–1803. [Google Scholar] [CrossRef] [PubMed]
- Graves, M.V.; Burbank, D.E.; Roth, R.; Heuser, J.; DeAngelis, P.L.; Van Etten, J.L. Hyaluronan synthesis in virus PBCV-1-infected chlorella-like green algae. Virology 1999, 257, 15–23. [Google Scholar] [CrossRef] [PubMed]
- DeAngelis, P.L. Hyaluronan synthases: fascinating glycosyltransferases from vertebrates, bacterial pathogens, and algal viruses. Cell. Mol. Life Sci. 1999, 56, 670–682. [Google Scholar] [CrossRef] [PubMed]
- DeAngelis, P.L. Evolution of glycosaminoglycans and their glycosyltransferases: Implications for the extracellular matrices of animals and the capsules of pathogenic bacteria. Anat. Rec. 2002, 268, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Landstein, D.; Graves, M.V.; Burbank, D.E.; DeAngelis, P.; Van Etten, J.L. Chlorella virus PBCV-1 encodes functional glutamine: fructose-6-phosphate amidotransferase and UDP-glucose dehydrogenase enzymes. Virology 1998, 250, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Duncan, G.; Agarkova, I.; Borodovsky, M.; Gurnon, J.; Kuo, A.; Lindquist, E.; Lucas, S.; Pangilinan, J.; Polle, J.; et al. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 2010, 22, 2943–2955. [Google Scholar] [CrossRef] [PubMed]
- Gooday, G.W.; Humphreys, A.M.; McIntosh, W.H. Roles of chitinases in fungal growth. In Chitin in nature and technology; Muzzarelli, R., Jeuniaux, C., Gooday, G.W., Eds.; Springer: Boston, MA, USA, 1986; pp. 83–91. [Google Scholar]
- Kapaun, E.; Loos, E.; Reisser, W. Cell wall composition of virus-sensitive symbiotic Chlorella species. Phytochemistry 1992, 31, 3103–3104. [Google Scholar] [CrossRef]
- Kawasaki, T.; Tanaka, M.; Fujie, M.; Usami, S.; Sakai, K.; Yamada, T. Chitin synthesis in chlorovirus CVK2-infected chlorella cells. Virology 2002, 302, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Kawasaki, T.; Yamada, T. Genetic rearrangements on the chlorovirus genome that switch between hyaluronan synthesis and chitin synthesis. Virology 2005, 342, 102–110. [Google Scholar]
- Yamada, T.; Kawasaki, T. Microbial synthesis of hyaluronan and chitin: New approaches. J. Biosci. Bioeng. 2005, 99, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Chase, T.E.; Nelson, J.A.; Burbank, D.E.; Van Etten, J.L. Mutual exclusion occurs in a Chlorella-like green alga inoculated with two viruses. J. Gen. Virol. 1989, 70, 1829–1836. [Google Scholar] [CrossRef] [PubMed]
- Greiner, T.; Frohns, F.; Kang, M.; Van Etten, J.L.; Käsmann, A.; Moroni, A.; Hertel, B.; Thiel, G. Chlorella viruses prevent multiple infections by depolarizing the host membranes. J. Gen. Virol. 2009, 90, 2033–2039. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.; Van Etten, J.L. Does extracellular hyaluronan and/or chitin confer an evolutionary advantage to the chloroviruses? Unpublished.
- Rakkhumkaew, N.; Kawasaki, T.; Fujie, M.; Yamada, T. Prolonged synthesis of hyaluronan by chlorella cells infected with chloroviruses. J. Biosci. Bioeng. 2013, 115, 527–531. [Google Scholar] [CrossRef] [PubMed]
- Tonetti, M.; Zanardi, D.; Gurnon, J.R.; Fruscione, F.; Armirotti, A.; Damonte, G.; Sturla, L.; De Flora, A.; Van Etten, J.L. Paramecium bursaria chlorella virus 1 encodes two enzymes involved in the biosynthesis of GDP-L-fucose and GDP-D-rhamnose. J. Biol. Chem. 2003, 278, 21559–21565. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Simala-Grant, J.L.; Taylor, D.E. Fucosylation in prokaryotes and eukaryotes. Glycobiology 2006, 16, 158r–184r. [Google Scholar] [CrossRef] [PubMed]
- De Castro, C.; Speciale, I.; Duncan, G.; Dunigan, D.D.; Agarkova, I.; Lanzetta, R.; Sturiale, L.; Palmigiano, A.; Garozzo, D.; Molinaro, A.; et al. N-linked glycans of chloroviruses sharing a core architecture without precedent. Angew. Chem. Int. Ed. 2016, 55, 654–658. [Google Scholar] [CrossRef] [PubMed]
- Speciale, I.; Agarkova, I.; Duncan, G.; Van Etten, J.L.; De Castro, C. Structure of the N-glycan from the chlorovirus NE-JV-1. A. Van. Leeuw. 2017. In press. [Google Scholar] [CrossRef] [PubMed]
- Rosano, C.; Zuccotti, S.; Sturla, L.; Fruscione, F.; Tonetti, M.; Bolognesi, M. Quaternary assembly and crystal structure of GDP-D-mannose 4,6 dehydratase from Paramecium bursaria chlorella virus. Biochem. Biophys. Res. Commun. 2006, 339, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Fruscione, F.; Sturla, L.; Duncan, G.; Van Etten, J.L.; Valbuzzi, P.; De Flora, A.; Di Zanni, E.; Tonetti, M. Differential role of NADP+ and NADPH in the activity and structure of GDP-D-mannose 4,6-dehydratase from two chlorella viruses. J. Biol. Chem. 2008, 283, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Chothi, M.P.; Duncan, G.A.; Armirotti, A.; Abergel, C.; Gurnon, J.R.; Van Etten, J.L.; Bernardi, C.; Damonte, G.; Tonetti, M. Identification of an L-rhamnose synthetic pathway in two nucleocytoplasmic large DNA viruses. J. Virol. 2010, 84, 8829–8838. [Google Scholar] [CrossRef] [PubMed]
- Doms, R.W.; Lamb, R.A.; Rose, J.K.; Helenius, A. Folding and assembly of viral membrane proteins. Virology 1993, 193, 545–562. [Google Scholar] [CrossRef] [PubMed]
- Olofsson, S.; Hansen, J.E. Host cell glycosylation of viral glycoproteins--a battlefield for host defence and viral resistance. Scand. J. Infect. Dis. 1998, 30, 435–440. [Google Scholar] [PubMed]
- Flint, S.J.; Racaniello, V.R.; Rail, G.F.; Skalka, A.M.; Enquist, L.W. Principles of Virology, 4th ed.; ASM Press: Washington, DC, USA, 2015; Vol. 1, p. 569. [Google Scholar]
- Vigerust, D.J.; Shepherd, V.L. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 2007, 15, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.N.; Li, Y.; Que, Q.; Bhattacharya, M.; Lane, L.C.; Chaney, W.G.; Van Etten, J.L. Evidence for virus-encoded glycosylation specificity. Proc. Natl. Acad. Sci. USA 1993, 90, 3840–3844. [Google Scholar] [CrossRef] [PubMed]
- De Castro, C.; Speciale, I.; Agarkova, I.; Molinaro, A.; Tonetti, M.; Duncan, G.A.; Van Etten, J.L. Glycan structures of chlorovirus PBCV-1 major capsid protein antigenic variants. Manuscript in preparation.
- De Castro, C.; Speciale, I.; Agarkova, I.; Molinaro, A.; Duncan, G.; Lowary, T.; Lee, S.; Jimenez-Barbero, J.; Tonetti, M.; Van Etten, J.L. Disclosure of the first viral encoded glycosyl transferase: Domain 1 of A064R gene of Paramecium bursaria chlorella virus encodes for a functional b-L-rhamnosyl transferase. Manuscript in preparation.
- Graves, M.V.; Bernadt, C.T.; Cerny, R.; Van Etten, J.L. Molecular and genetic evidence for a virus-encoded glycosyltransferase involved in protein glycosylation. Virology 2001, 285, 332–345. [Google Scholar] [CrossRef] [PubMed]
- Van Etten, J.L.; Burbank, D.E.; Xia, Y.; Meints, R.H. Growth cycle of a virus, PBCV- 1, that infects chlorella-like algae. Virology 1983, 126, 117–125. [Google Scholar] [CrossRef]
- Nandhagopal, N.; Simpson, A.A.; Gurnon, J.R.; Yan, X.; Baker, T.S.; Graves, M.V.; Van Etten, J.L.; Rossmann, M.G. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl. Acad. Sci. USA 2002, 99, 14758–14763. [Google Scholar] [CrossRef] [PubMed]
- De Castro, C.; Speciale, I.; Molinaro, A.; Van Etten, J.L.; Klose, T.; Rossmann, M.G. Major capsid protein of chlorovirus PBCV-1 structure combining X-ray and carbohydrate molecular modeling approaches. Proc. Natl. Acad. Sci. USA. In revision.
- Zielinska, D.F.; Gnad, F.; Wisniewski, J.R.; Mann, M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 2010, 141, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, F.; Aebi, M. Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 2011, 21, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Abu-Qarn, M.; Yurist-Doutsch, S.; Giordano, A.; Trauner, A.; Morris, H.R.; Hitchen, P.; Medalia, O.; Dell, A.; Eichler, J. Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. J. Mol. Biol. 2007, 374, 1224–1236. [Google Scholar] [CrossRef] [PubMed]
- Chaney, W.G.; Van Etten, J.L. Inability to cleave glycans from chlorovirus PBCV-1 major capsid protein by traditional glycosidases. Unpublished.
- De Castro, C.; Molinaro, A.; Piacente, F.; Gurnon, J.R.; Sturiale, L.; Palmigiano, A.; Lanzetta, R.; Parrilli, M.; Garozzo, D.; Tonetti, M.G.; et al. Structure of N-linked oligosaccharides attached to chlorovirus PBCV-1 major capsid protein reveals unusual class of complex N-glycans. Proc. Natl. Acad. Sci. USA 2013, 110, 13956–13960. [Google Scholar] [CrossRef] [PubMed]
- Wieland, F.; Heitzer, R.; Schaefer, W. Asparaginylglucose: novel type of carbohydrate linkage. Proc. Natl. Acad. Sci. USA 1983, 80, 5470–5474. [Google Scholar] [CrossRef] [PubMed]
- Mengele, R.; Sumper, M. Drastic differences in glycosylation of related S-layer glycoproteins from moderate and extreme halophiles. J. Biol. Chem. 1992, 267, 8182–8185. [Google Scholar] [PubMed]
- Schreiner, R.; Schnabel, E.; Wieland, F. Novel N-glycosylation in eukaryotes: laminin contains the linkage unit beta-glucosylasparagine. J. Cell. Biol. 1994, 124, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Gross, J.; Grass, S.; Davis, A.E.; Gilmore-Erdmann, P.; Townsend, R.R.; St Geme, J.W. The Haemophilus influenzae HMW1 adhesin is a glycoprotein with an unusual N-linked carbohydrate modification. J. Biol. Chem. 2008, 283, 26010–26015. [Google Scholar] [CrossRef] [PubMed]
- Que, Q.; Li, Y.; Wang, I.N.; Lane, L.C.; Chaney, W.G.; Van Etten, J.L. Protein glycosylation and myristylation in chlorella virus PBCV-1 and its antigenic variants. Virology 1994, 203, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Chuchird, N.; Nishida, K.; Kawasaki, T.; Fujie, M.; Usami, S.; Yamada, T. A variable region on the chlorovirus CVK2 genome contains five copies of the gene for Vp260, a viral-surface glycoprotein. Virology 2002, 295, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Yanai-Balser, G.M.; Duncan, G.A.; Eudy, J.D.; Wang, D.; Li, X.; Agarkova, I.V.; Dunigan, D.D.; Van Etten, J.L. Microarray analysis of chlorella virus PBCV-1 transcription. J. Virol. 2010, 84, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.P.; Schultz, J.; Mlodzik, M.; Bork, P. Secreted fringe-like signaling molecules may be glycosyltransferases. Cell 1997, 9–11. [Google Scholar] [CrossRef]
- Bruckner, K.; Perez, L.; Clausen, H.; Cohen, S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature 2000, 406, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xiang, Y.; Van Etten, J.L.; Rossmann, M.G. Structure and function of a chlorella virus encoded glycosyltransferase. Structure 2007, 15, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Baxa, U.; Zhang, Y.; Steven, A.C.; Lewis, G.L.; Van Etten, J.L.; Rossmann, M.G. Crystal structure of a virus-encoded putative glycosyltransferase. J. Virol. 2010, 84, 12265–12273. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, P.M.; Deleury, E.; Davies, G.J.; Henrissat, B. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 2003, 328, 307–317. [Google Scholar] [CrossRef]
- Lairson, L.L.; Henrissat, B.; Davies, G.J.; Withers, S.G. Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 2008, 77, 521–555. [Google Scholar] [CrossRef] [PubMed]
- Van Etten, J.L.; Gurnon, J.R.; Yanai-Balser, G.M.; Dunigan, D.D.; Graves, M.V. Chlorella viruses encode most, if not all, of the machinery to glycosylate their glycoproteins independent of the endoplasmic reticulum and Golgi. Biochim. Biophys. Acta 2010, 1800, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xiang, Y.; Dunigan, D.D.; Klose, T.; Chipman, P.R.; Van Etten, J.L.; Rossmann, M.G. Three-dimensional structure and function of the Paramecium bursaria chlorella virus capsid. Proc. Natl. Acad. Sci. USA 2011, 108, 14837–14842. [Google Scholar] [CrossRef] [PubMed]
- Meints, R.H.; Lee, K.; Burbank, D.E.; Van Etten, J.L. Infection of a chlorella-like alga with the virus, PBCV-1: Ultrastructural studies. Virology 1984, 138, 341–346. [Google Scholar] [CrossRef]
- Milrot, E.; Shimoni, E.; Dadosh, T.; Unger, T.; Van Etten, J.L.; Minsky, A. Bacteriophage-like infection of the large eukaryote-infecting Paramecium bursaria chlorella virus-1. PLoS Pathogens, Submitted.
- Frohns, F.; Käsmann, A.; Kramer, D.; Schäfer, B.; Mehmel, M.; Kang, M.; Van Etten, J.L.; Gazzarrini, S.; Moroni, A.; Thiel, G. Potassium ion channels of chlorella viruses cause rapid depolarization of host cells during infection. J. Virol. 2006, 80, 2437–2444. [Google Scholar] [CrossRef] [PubMed]
- Neupärtl, M.; Meyer, C.; Woll, I.; Frohns, F.; Kang, M.; Van Etten, J.L.; Kramer, D.; Hertel, B.; Moroni, A.; Thiel, G. Chlorella viruses evoke a rapid release of K+ from host cells during early phase of infection. Virology 2008, 372, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Adams, B.; Gurnon, J.; Ye, Y.; Van Etten, J.L. Characterization of two chitinase genes and one chitosanase gene encoded by chlorella virus PBCV-1. Virology 1999, 263, 376–387. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, S.; Ishihara, M.; Fujie, M.; Usami, S.; Yamada, T. Expression of a chitinase gene and lysis of the host cell wall during Chlorella virus CVK2 infection. Virology 1999, 260, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Hiramatsu, S.; Songsri, P.; Fujie, M. Alternative expression of a chitosanase gene produces two different proteins in cells infected with chlorella virus CVK2. Virology 1997, 230, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Gurnon, J.R.; Adams, B.J.; Graves, M.V.; Van Etten, J.L. Characterization of a β-1,3-glucanase encoded by chlorella virus PBCV-1. Virology 2000, 276, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Suda, K.; Tanji, Y.; Hori, K.; Unno, H. Evidence for a novel chlorella virus-encoded alginate lyase. FEMS Microbiol. Lett. 1999, 180, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, I.; Onimatsu, H.; Fujie, M.; Usami, S.; Yamada, T. vAL-1, a novel polysaccharide lyase encoded by chlorovirus CVK2. FEBS Lett. 2004, 559, 51–56. [Google Scholar] [CrossRef]
- Ogura, K.; Yamasaki, M.; Yamada, T.; Mikami, B.; Hashimoto, W.; Murata, K. Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action. J. Biol. Chem. 2009, 284, 35572–35579. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, I.; Hiramatsu, S.; Murakami, D.; Fujie, M.; Usami, S.; Yamada, T. Algal-lytic activities encoded by Chlorella virus CVK2. Virology 2000, 277, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Meints, R.H.; Burbank, D.E.; Van Etten, J.L.; Lamport, D.T. Properties of the chlorella receptor for the virus PBCV-1. Virology 1988, 164, 15–21. [Google Scholar] [CrossRef]
- Agarkova, I.; Hertel, B.; Zhang, X.; Lane, L.; Tchourbanov, A.; Dunigan, D.D.; Thiel, G.; Rossmann, M.G.; Van Etten, J.L. Dynamic attachment of chlorovirus PBCV-1 to Chlorella variabilis. Virology 2014, 466–467, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Agarkova, I.V.; Lane, L.C.; Dunigan, D.D.; Quispe, C.; Duncan, G.A.; Milrot, E.; Minsky, A.; Esmael, A.; Van Etten, J.L. Identification of the chlorovirus PBCV-1 enzyme involved in degrading the host cell wall during virus infection. Manuscript in preparation.
- Furuta, M.; Schrader, J.O.; Schrader, H.S.; Kokjohn, T.A.; Nyaga, S.; McCullough, A.K.; Lloyd, R.S.; Burbank, D.E.; Landstein, D.; Lane, L.; et al. Chlorella virus PBCV-1 encodes a homolog of the bacteriophage T4 UV damage repair gene denV. Appl. Environ. Microbiol. 1997, 63, 1551–1556. [Google Scholar] [PubMed]
- McCullough, A.K.; Romberg, M.T.; Nyaga, S.; Wei, Y.; Wood, T.G.; Taylor, J.S.; Van Etten, J.L.; Dodson, M.L.; Lloyd, R.S. Characterization of a novel cis-syn and trans-syn-II pyrimidine dimer glycosylase/AP lyase from a eukaryotic algal virus, Paramecium bursaria chlorella virus-1. J. Biol. Chem. 1998, 273, 13136–13142. [Google Scholar] [CrossRef] [PubMed]
- Piacente, F.; Gaglianone, M.; Laugieri, M.E.; Tonetti, M.G. The autonomous glycosylation of large DNA viruses. Int. J. Mol. Sci. 2015, 16, 29315–29328. [Google Scholar] [CrossRef] [PubMed]
- Moreau, H.; Piganeau, G.; Desdevises, Y.; Cooke, R.; Derelle, E.; Grimsley, N. Marine prasinovirus genomes show low evolutionary divergence and acquisition of protein metabolism genes by horizontal gene transfer. J. Virol. 2010, 84, 12555–12563. [Google Scholar] [CrossRef] [PubMed]
- Santini, S.; Jeudy, S.; Bartoli, J.; Poirot, O.; Lescot, M.; Abergel, C.; Barbe, V.; Wommack, K.E.; Noordeloos, A.A.; Brussaard, C.P.; et al. Genome of Phaeocystis globosa virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes. Proc. Natl. Acad. Sci. USA 2013, 110, 10800–10805. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.G.; Allen, M.J.; Wilson, W.H.; Suttle, C.A. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc. Natl. Acad. Sci. USA 2010, 107, 19508–19513. [Google Scholar] [CrossRef] [PubMed]
- Philippe, N.; Legendre, M.; Doutre, G.; Coute, Y.; Poirot, O.; Lescot, M.; Arslan, D.; Seltzer, V.; Bertaux, L.; Bruley, C.; et al. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 2013, 341, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Legendre, M.; Bartoli, J.; Shmakova, L.; Jeudy, S.; Labadie, K.; Adrait, A.; Lescot, M.; Poirot, O.; Bertaux, L.; Bruley, C.; et al. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc. Natl. Acad. Sci. USA 2014, 111, 4274–4279. [Google Scholar] [CrossRef] [PubMed]
- Legendre, M.; Lartigue, A.; Bertaux, L.; Jeudy, S.; Bartoli, J.; Lescot, M.; Alempic, J.M.; Ramus, C.; Bruley, C.; Labadie, K.; et al. In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc. Natl. Acad. Sci. USA 2015, 112, E5327–E5335. [Google Scholar] [CrossRef] [PubMed]
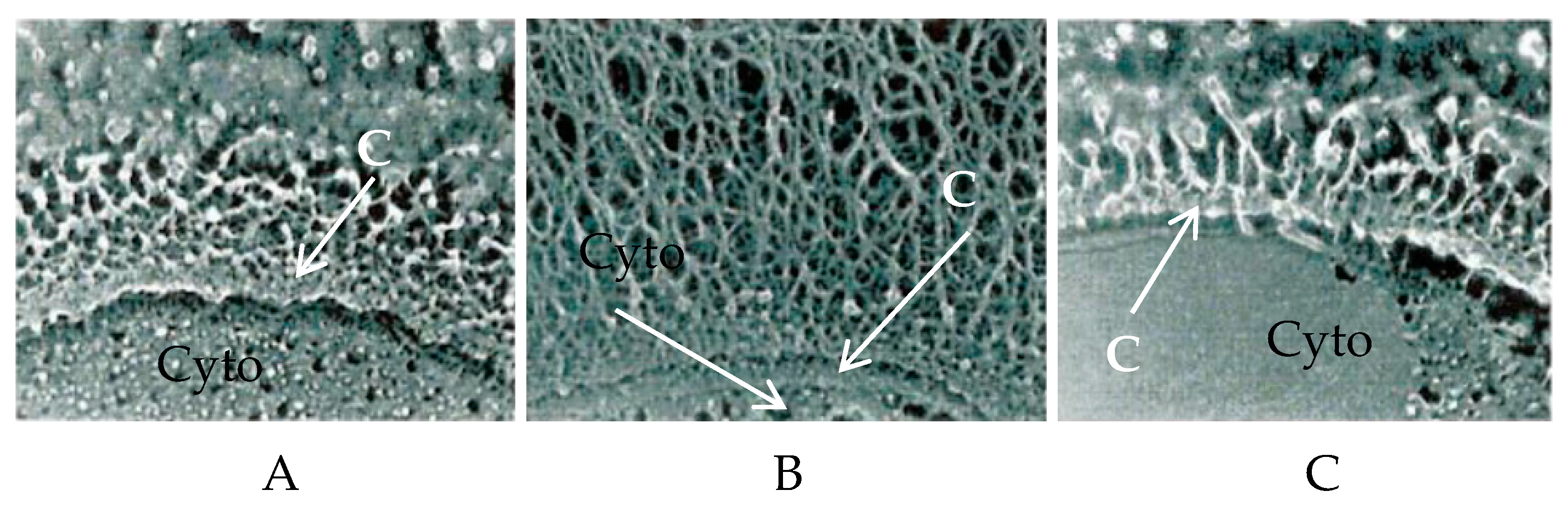
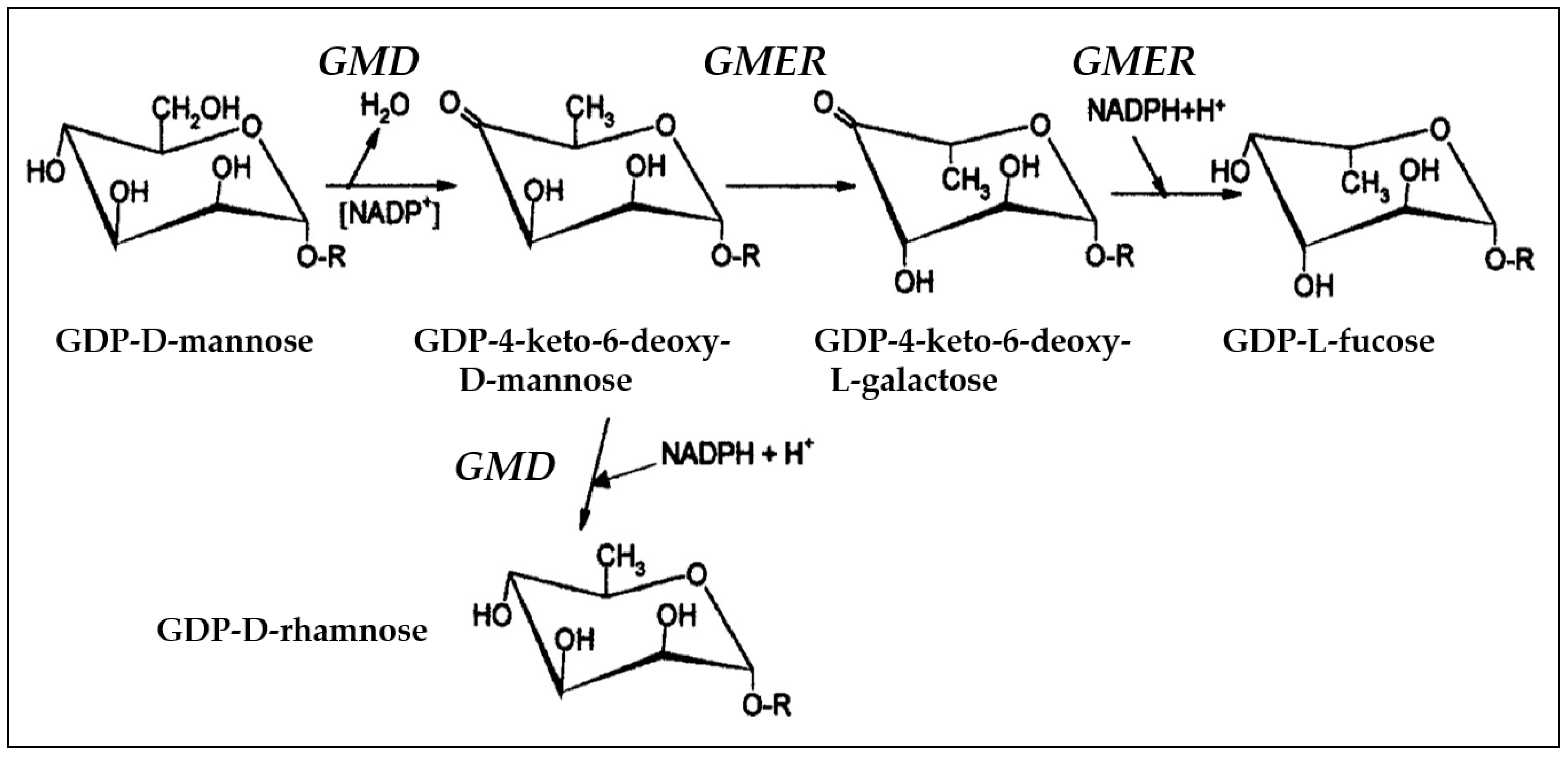
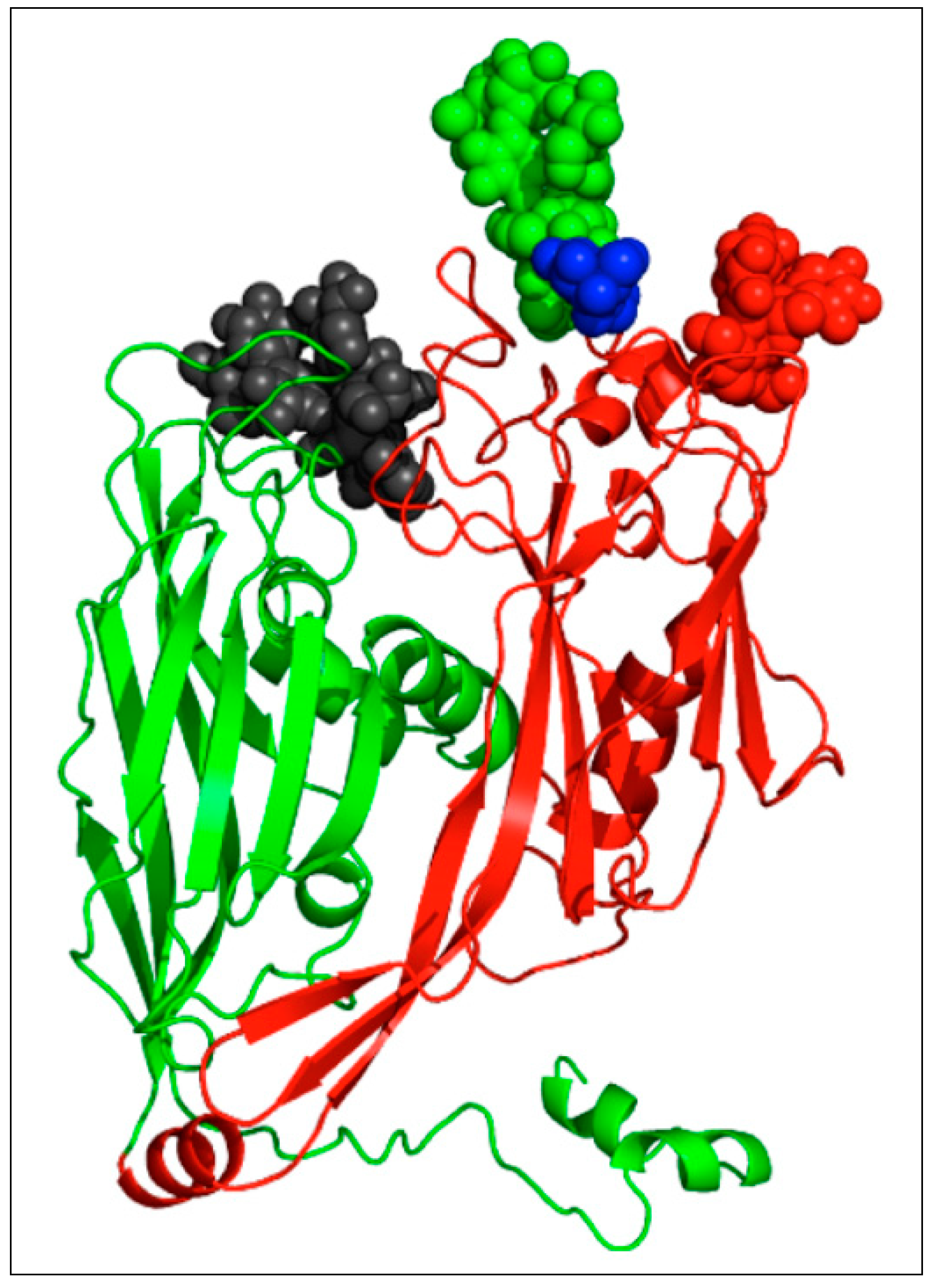
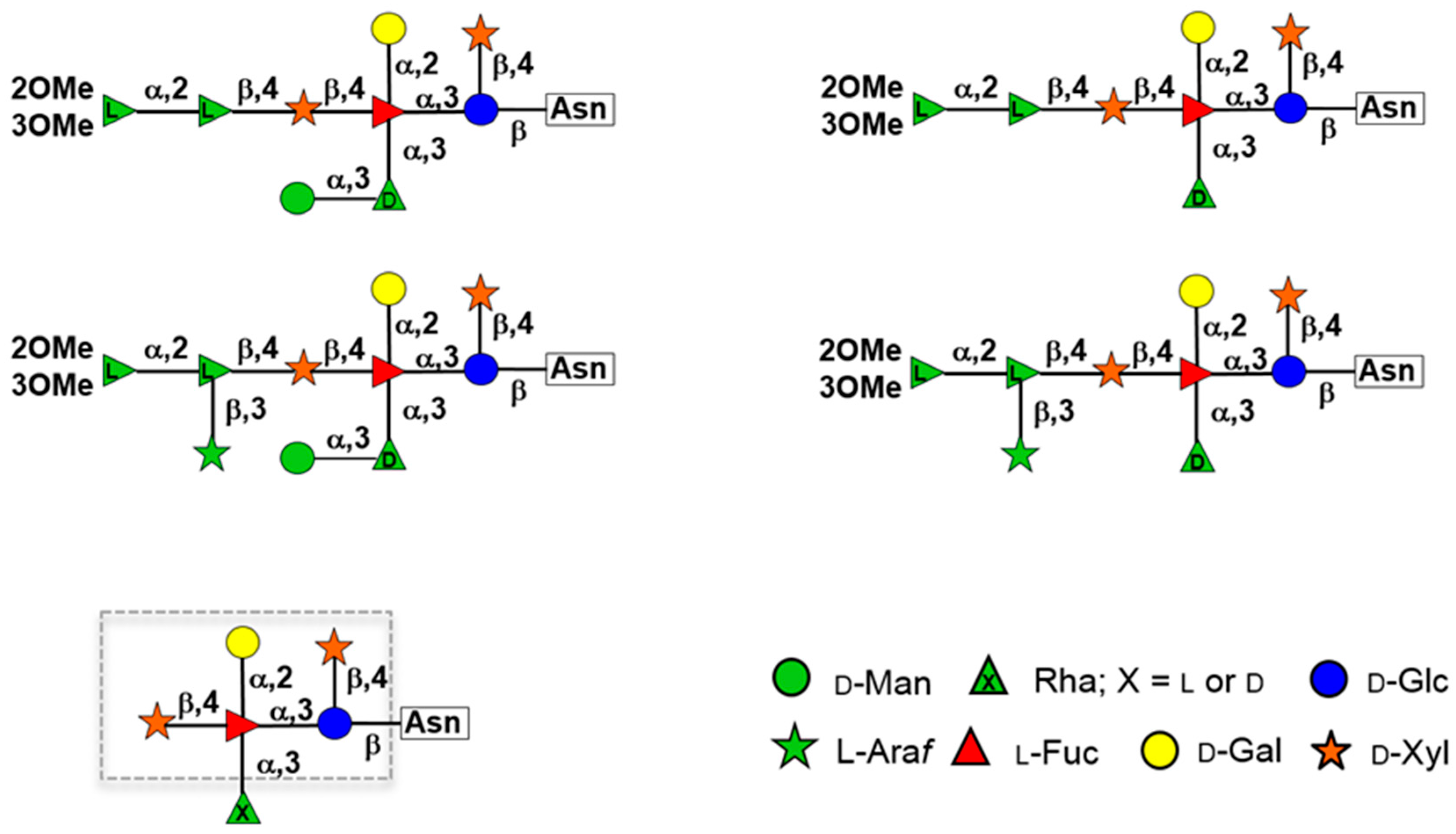
| Host | Viruses | HAS 1 | CHS 2 | CBP 3 |
|---|---|---|---|---|
| NC64A | AN69C | 390R | 395L, 438L | |
| AR158 | C418R | C423L, C475L | ||
| CviK1 | 102R | 365R | 370L, 414L | |
| CvsA1 | 375R | 380L, 427L | ||
| IL-3A | 386L, 432L | |||
| IL-5-2s1 | 134L | 503L, 562L | ||
| KS-1B | 314L, 360L | |||
| MA-1D | 485L | 362R | 367L, 472L | |
| MA-1E | 113L | 407L, 451L | ||
| NE-JV-4 | 390L | |||
| NY-2B | 542L, 484L | |||
| NY2A | B139R, B472R | B480L | ||
| NYs-1 | 137R | 360R, 495L, 555L | ||
| PBCV-1 | a A098R | A333L, A348Dl, A383R | ||
| SYN | OSY-NE-5 | 038R | 167L, 184L | |
| Pbi | AP110A | 152L, 175R | 828R | |
| CVA-1 | 150L, 169R | 834R | ||
| CVB-1 | 177L | 791R | ||
| CVG-1 | 146R | 792R | ||
| CVM-1 | 165L, 186R | 832R | ||
| CVR-1 | 838R | |||
| CZ-2 | 798R | |||
| Can18-4 | 163R | 839R | ||
| FR483 | N124R | N690R | ||
| FR5L | 151L | 797R | ||
| MT325 | M128R | M701R | ||
| NE-JV-1 | 278R, 282R | 734L | ||
| NW665 | 133R | 821R | ||
| OR0704.2.2 | 116R | 804R | ||
| SAG | ATCV-1 | Z734R | ||
| Br0604L | 834R | 431R | ||
| Can0610SP | 438R, 442R | |||
| Canal-1 | 746R | 405R | ||
| GM0701 | 852R | 436R | ||
| MN0810 | 087R, 900R | 466R, 468R, 531L | ||
| MO0605SPH | 435R | |||
| NE-JV-2 | 462R | |||
| NE-jv-3 | 431R | |||
| NTS-1 | 893R | 461R, 463R, 529L | ||
| OR0704.3 | 431R | |||
| TN603 | 869R | 425R | ||
| W10606 | 457R |
| Host | Viruses | GFAT 1 | UDP-GlcDH 2 | GMD 3 | GMER 4 | UGD 5 | AT 6 | D-LD 7 | ADP-RGH 8 | FRD 9 |
|---|---|---|---|---|---|---|---|---|---|---|
| NC64A | AN69C | 109R | 384R, 487R | 129R | 334L | 739L | 055R | |||
| AR158 | C132R | C413R, C729L | C155R | C344L | C767L | |||||
| CviK1 | 105R | 359R, 662L | 122R | 312L | 742L | 055R | ||||
| CvsA1 | 064R | 368R | 083R | 321L | 716L | 131R | ||||
| IL-3A | 104R | 375R, 685L | 126R | 326L | 726L | 051R | ||||
| IL-5-2s1 | 130R | 492R, 858L | 106L | 417L | 896L | |||||
| KS-1B | 066R | 261L | 643L | |||||||
| MA-1D | 481L | 355R, 838L | 456L | 284L | 872L | |||||
| MA-1E | 116R | 396R | 134R | 355L | 806L | 045R | ||||
| NE-JV-4 | 131R | 340L | 740L | 064R | ||||||
| NY-2B | 473R, 836L | 185R | 408L | 881L | ||||||
| NY2A | B143R | B465R | B163R | B395L | B853L | |||||
| NYs-1 | 143R | 483R, 846L | 167R | 404L | 879L | |||||
| PBCV-1 | a A100R | a A609L | a A118R | a A295L | A654L | A053R | ||||
| SYN | OSY-NE-5 | 039R | 045R | 139L | 340L | 015R | 308R | |||
| Pbi | AP110A | 071R | 146L | 893R | 053L | |||||
| CVA-1 | 056R | 144L | 900R | 040L | 205R | |||||
| CVB-1 | 071R | 172L | 856R | 056L | 221R | |||||
| CVG-1 | 050R | 812L | 857R | |||||||
| CVM-1 | 069R | 159L | 893R | 052L | 222R | |||||
| CVR-1 | 062R | 151L | 906R | 046L | 210R | |||||
| CZ-2 | 059R | 718L | 865R | 048L | 917L | |||||
| Can18-4 | 061R | 859L | 908R | 048L | 212R | |||||
| FR483 | N035R | N712L | N747R | N170R | ||||||
| FR5L | 087R | 145L | 863R | 076L | ||||||
| MT325 | M036R, M037R | M719L | M758R | M026L | ||||||
| NE-JV-1 | 081R | 291R | 861R | 810L | ||||||
| NW665 | 046R | 846L | 889R | 189R | ||||||
| OR0704.2.2 | 062R | 722L | 862R | 051L | ||||||
| SAG | ATCV-1 | Z571L | a Z804L | Z282L | Z544R | Z147L | Z295L | |||
| Br0604L | 667L, 839R | 934L | 332L | 631R | 173L | 350L | ||||
| Can0610SP | 687L | 965L | 338L | 658R | 170L | 355L | ||||
| Canal-1 | 605L, 751R | 847L | 329L | 576R | 188L | 343L | 898L | 886R | ||
| GM0701 | 664L, 856R | 954L | 337L | 629R | 180L | 354L | ||||
| MN0810 | 887L | 720L, 904R | 992L | 365L | 689R | 204L | 379L | |||
| MO0605SPH | 656L | 897L | 341L | 625R | 181L | 359L | 943R | |||
| NE-JV-2 | 708L | 981L | 367L | 672R | 186L | 383L | ||||
| NE-JV-3 | 679L | 935L | 332L | 648R | 175L | 347L | 981R | |||
| NTS-1 | 714L, 898R | 1012L | 378L | 681R | 188L | 391L | ||||
| OR0704.3 | 676L | 960L | 335L | 639R | 179L | 354L | ||||
| TN603 | 659L, 873R | 966L | 326L | 625R | 179L | 342L | ||||
| W10606 | 679L | 916L | 360L | 651R | 185L | 375L | 962R |
| Antisera Classes | Predicted MW (kDa) c | SDS-PAGE Estimates (kDa) d | |
|---|---|---|---|
| Class a | Label b | ||
| + | Wild-type | 54.1 | 54 |
| F | CME6 | 54 | not determined |
| A | P91 | 52.8 | 53 |
| E | EPA-15 | 52.8 | not determined |
| B | EPA-2 | 51.6 | 52 |
| C | E1L3 | 51.1 | 51 |
| D | P1L6 | 50.5 | 50.5 |
| Host | Viruses | EXT 1 | GT-A 2 | GT-GT4 3 | GT-GTA 4 | CESA CelA-Like 5 | CSCS-2 6 | GT 7 | GT17 8 |
|---|---|---|---|---|---|---|---|---|---|
| NC64A | AN69C | 078L | 123R | 559R | 065R | 104R | 255R | ||
| AR158 | C093L | C150R | C661L | C265R | C559R | ||||
| CviK1 | 080L | 117R | 594L | 518L | 242R | ||||
| CvsA1 | 039L | 077R | 611L | 535L | 247R | ||||
| IL-3A | 071L | 120R | 606L | 060R | 099R | 240R | |||
| IL-5-2s1 | 175R | 109L | 773L | 313R | 649R | ||||
| KS-1B | 024L | 060R | 528L | 009R | 046L | 170R | |||
| MA-1D | 531R | 4549L | 753L | 194R | 637R | ||||
| MA-1E | 533R | 128R | 702L | 626L | 281R, 210R | ||||
| NE-JV-4 | 085L | 124R | 631L | 074R | 108L | 250R | |||
| NY-2B | 116L | 180R | 754L | 160R | 323R | 633R | |||
| NY2A | 107L | B159R | B736L | B618R | |||||
| NYs-1 | 098L | 162R | 760L | 187R | 641R | ||||
| PBCV-1 | A075L | A111/114R | A546L | A064R | A473L | A219/222/226R | |||
| SYN | OSY-NE-5 | 025L | 044R | 283L | 097L | ||||
| Pbi | AP110A | 013L | 548R | 226R | 970R | ||||
| CVA-1 | 016L | 532R | 220R | 971R | |||||
| CVB-1 | 025L | 538R | 232R | 811L | 918R | ||||
| CVG-1 | 019L | 520R | 217R | 815L | 920R | ||||
| CVM-1 | 022L | 550R | 237R | 953R | |||||
| CVR-1 | 020L | 545R | 225R | 977R | |||||
| CZ-2 | 012L | 532R | 380L | 822L | 932R | ||||
| Can18-4 | 020L | 557R | 229R | 862L | 971R | ||||
| FR483 | N012L | N472R | N191R | N715L | N805R | ||||
| FR5L | 046L | 537R | 819L | 926R | |||||
| MT325 | M009L | M467R | M186R | M721L | M813R | ||||
| NE-JV-1 | 079L | 464L | 801R | 930R | |||||
| NW665 | 015L | 532R | 849L | 955R | |||||
| OR0704.2.2 | 017L | 549R | 382L | 823L | 923R | ||||
| SAG | ATCV-1 | Z830R | Z120R | Z667L | Z178L, Z823R, Z417L | Z425R | Z347R | ||
| Br0604L | 959R | 137R | 225L, 952R, 483L | 489R | 399R | ||||
| Can0610SP | 1007R | 140R | 789L | 210L, 1002R, 978L, 487L | 495R | 407R | |||
| Canal-1 | 874R | 141R | 164L | 871R | 447R | 380R | |||
| GM0701 | 977R | 141R | 752L | 228L, 975R, 486L | 493R | 405R | |||
| MN0810 | 1009R | 165R | 244L | 424R | |||||
| MO0605SPH | 932R | 138R | 164L | 230L, 926R, 479L | 488R | 412R | |||
| NE-JV-2 | 1020R | 150R | 804L | 1015R, 992L, 516L | 523R | 438R | |||
| NE-JV-3 | 970R | 145R | 778L | 215L, 210L, 964R, 484L | 493R | 407R | |||
| NTS-1 | 1044R | 156R | 1016L, 516L | 441R | |||||
| OR0704.3 | 1006R | 146R | 944L | 1001R, 972L, 485L, 219L | 493R | 404R | |||
| TN603 | 991R | 141R | 226L, 986R, 475L | 483R | 400R | ||||
| WI0606 | 951R | 141R | 233L, 945R, 506L | 514R | 430R |
| Host | Viruses | CHI 1 | CHIS 2 | GUN 3 | BCHIL 4 | Lysin 5 | GH 6 | CD 7 | ALGL 8 |
|---|---|---|---|---|---|---|---|---|---|
| NC64A | AN69C | 297R | 331L | 102L | 204R | 540R | 387L | 250L | |
| AR158 | C342L | C126L | C220R | C681L | C415L | C263L | |||
| CviK1 | 309L | 99L | 194R | 610L | 362L | 237L | |||
| CvsA1 | 318L | 058L | 200R | 627L | 371L, 373L | 243L | |||
| IL-3A | 285R | 323L | 097L | 196R | 624L | 378L | 235L | ||
| IL-5-2s1 | 373R | 415L | 140R | 262R | 796L | 495L | 310L | ||
| KS-1B | 211R | 257L | 048R | 132R | 546L | 166L | |||
| MA-1D | 241R, 242R | 283L | 491R | 147R | 777L | 359L | 191L | ||
| MA-1E | 352L | 114R | 194R, 229R | 717L | 399L | 277L | |||
| NE-JV-4 | 298R | 337L | 110R | 205R | 648L | 246L | |||
| NY-2B | 367R | 406L | 158L | 270R | 777L | 476L | 319L | ||
| NY2A | B393L | B137L | B239R | B756L | B469L | B288L | |||
| NYs-1 | 360R | 403L | 134L | 251R | 487L | ||||
| PBCV-1 | a A260R | a A292L | a A094L | a A181/182R | a A561L | a A215L | |||
| SYN | OSY-NE-5 | 119R | 138L | 037L | 117L | 290L | 099R | ||
| Pbi | AP110A | 106R | 122R | 174L | 942R | 306R | 158L | 338R | |
| CVA-1 | 102R | 114R | 167L | 942R | 297R | 156L | 327R | ||
| CVB-1 | 129R | 143R | 890R | 310R | 183L | 342R | |||
| CVG-1 | 105R | 113R | 142L | 896R | 303R | 329R | |||
| CVM-1 | 122R | 133R | 184L | 927R | 330R | 171L | 360R | ||
| CVR-1 | 109R | 121R | 174L | 948R | 163L | 335R | |||
| CZ-2 | 070R | 079R | 114L | 902R | 263R | 293R | |||
| Can18-4 | 113R | 121R | 160L | 944R | 316R | 349R | |||
| FR483 | N087R | N119L | N779R | N262R | N293R | ||||
| FR5L | 098R | 106R | 897R | 300R | 157L | 336R | |||
| MT325 | M085R | M091R | M124L | M791R | M258R | M289R | |||
| NE-JV-1 | 328L | 218L | 275L | 088R | 592L | 050L | 269L | 472L | |
| NW665 | 088R | 128L | 921R | 281R | 315R | ||||
| OR0704.2.2 | 074R | 082R | 113L | 895R | 257R | 292R | |||
| SAG | ATCV-1 | Z780L | Z204R | Z819L | Z814L | Z511L | Z771L | ||
| Br0604L | 253R, 254R | 950L | 902R, 942L | 518L | 832L | 895L | |||
| Can0610SP | 244R | 985R, 1001L | 936R, 996R | 613L | 919L | ||||
| Canal-1 | 815L | 242R | 866L | 138L, 855L | 546L | 743L | 806L | ||
| GM0701 | 251R | 972L | 917R, 963L | 526L | 846L | 910L | |||
| MN0810 | 271R | 082L | 162L, 1003L | 659L | 896L | 959L | |||
| MO0605SPH | 871L | 257R | 920L | 911L | 588L | 862L | |||
| NE-JV-2 | 950L | 258R | 1000R, 1013L | 1008R | 634L | 932L | |||
| NE-JV-3 | 907L | 244R | 957L | 948L | 603L | 897L | |||
| NTS-1 | 973L | 276R | 1024R | 1039R | 556L | 886L | 961L | ||
| OR0704.3 | 251R | 979R, 999L | 918R, 994R | 604L | 910L | ||||
| TN603 | 253R | 983L | 939R, 977L | 520L | 867L | 931L | |||
| WI0606 | 889L | 262R | 938L | 929L | 612L | 878L |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van Etten, J.L.; Agarkova, I.; Dunigan, D.D.; Tonetti, M.; De Castro, C.; Duncan, G.A. Chloroviruses Have a Sweet Tooth. Viruses 2017, 9, 88. https://doi.org/10.3390/v9040088
Van Etten JL, Agarkova I, Dunigan DD, Tonetti M, De Castro C, Duncan GA. Chloroviruses Have a Sweet Tooth. Viruses. 2017; 9(4):88. https://doi.org/10.3390/v9040088
Chicago/Turabian StyleVan Etten, James L., Irina Agarkova, David D. Dunigan, Michela Tonetti, Christina De Castro, and Garry A. Duncan. 2017. "Chloroviruses Have a Sweet Tooth" Viruses 9, no. 4: 88. https://doi.org/10.3390/v9040088








