Poxvirus Host Range Genes and Virus–Host Spectrum: A Critical Review
Abstract
:1. Introduction
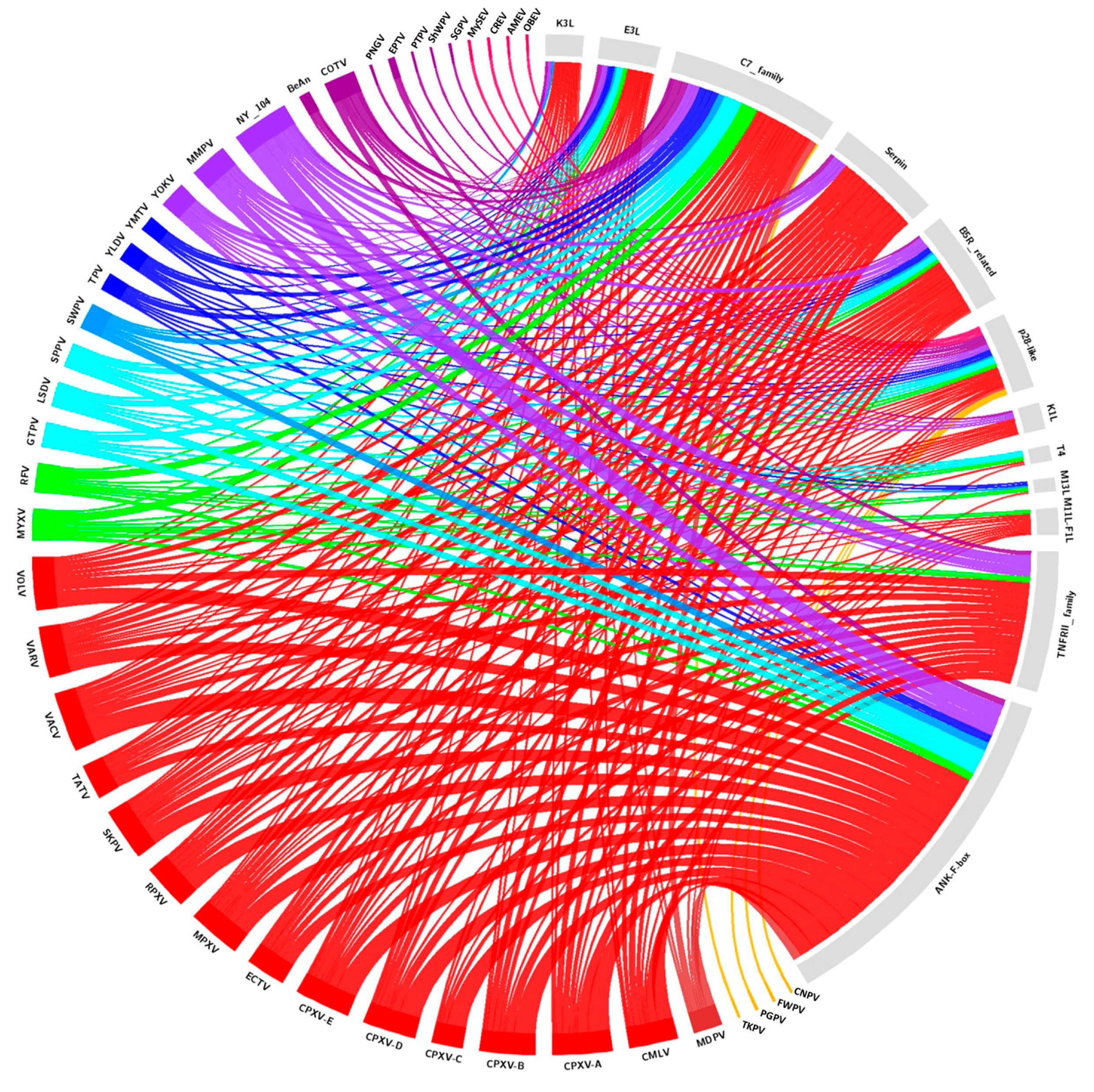
2. Diversity and Abundance of Host Range Factors Are Not Proportional to the Diversity of Hosts in Poxviridae
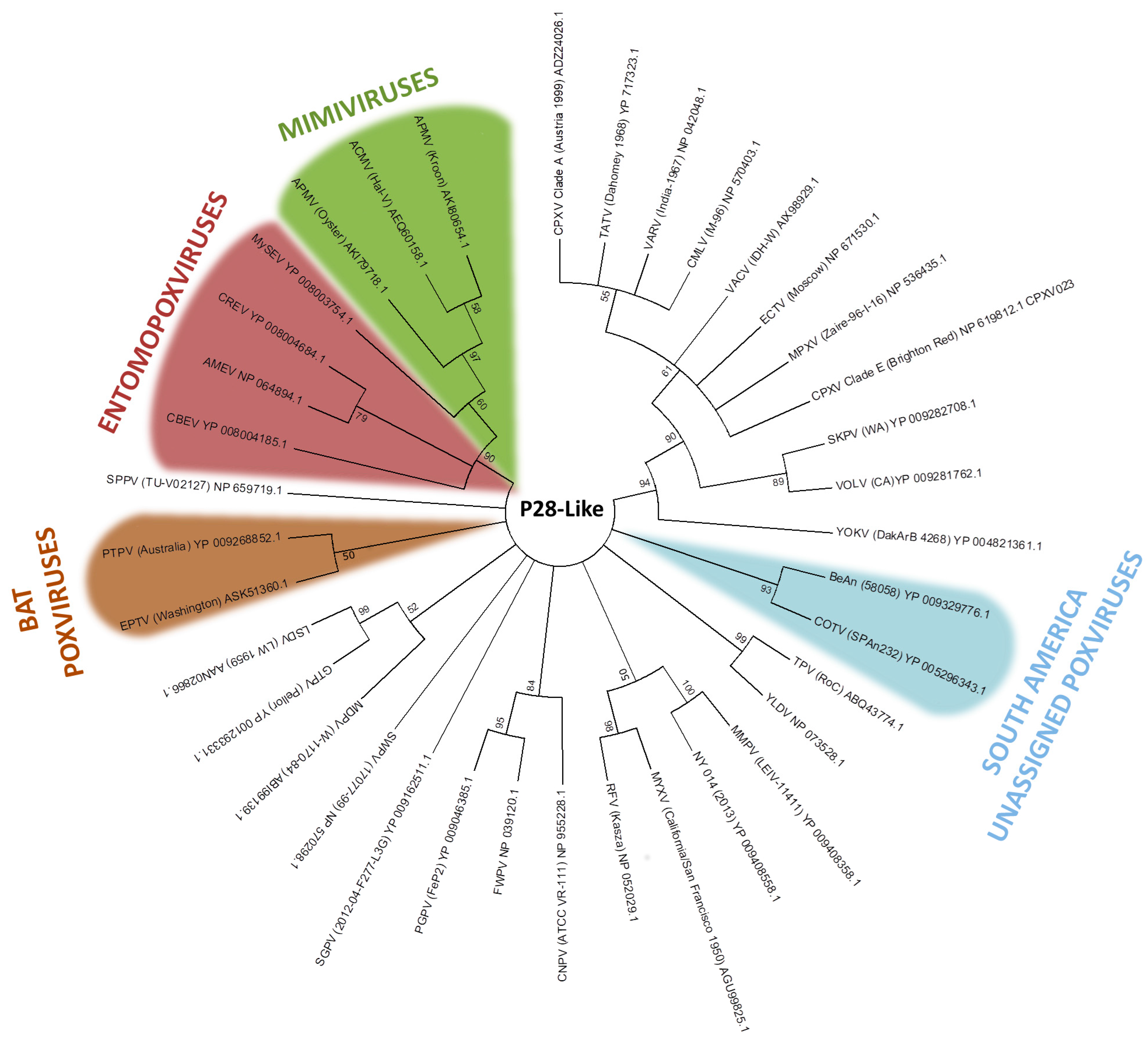
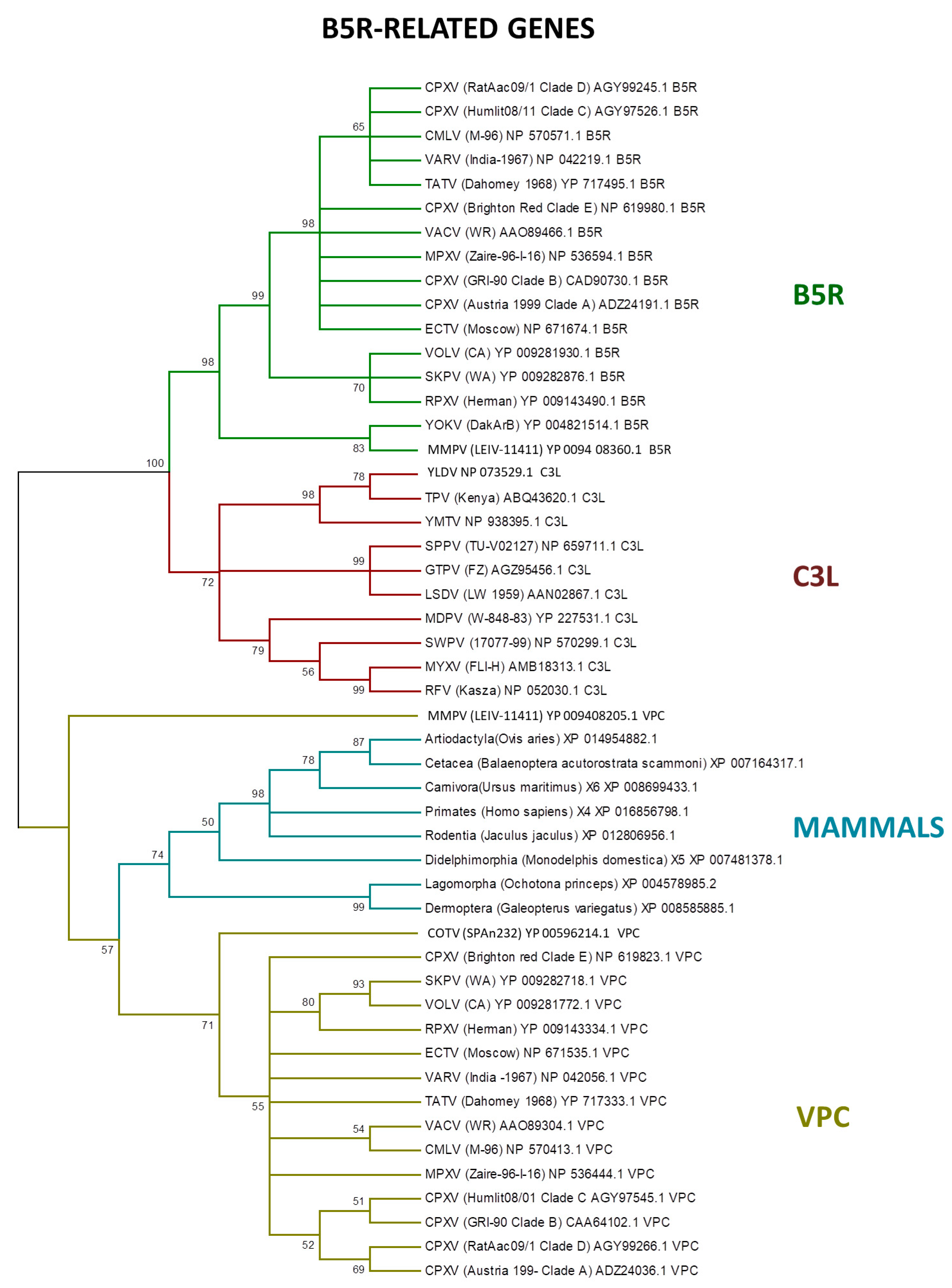
3. Host Range Genes: An Evolutionary Perspective
4. Future Directions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- International Committee on Taxonomy of Viruses—Taxonomy. Available online: https://talk.ictvonline.org/taxonomy/w/ictv-taxonomy (accessed on 1 July 2017).
- Fenner, F.; Henderson, D.A.; Arita, I.; Jezek, Z.; Ladnyi, I.D. Smallpox and Its Eradication, 1st ed.; World Health Organization: Geneva, Switzerland, 1988; pp. 1371–1409. [Google Scholar]
- Thèves, C.; Biagini, P.; Crubézy, E. The rediscovery of smallpox. Clin. Microbiol. Infect. 2014, 20, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Henderson, D.A. The eradication of smallpox—An overview of the past, present, and future. Vaccine 2011, 29, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Essbauer, S.; Pfeffer, M.; Meyer, H. Zoonotic poxviruses. Vet. Microbiol. 2010, 140, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Kroon, E.G.; Mota, B.E.F.; Abrahão, J.S.; da Fonseca, F.G.; de Souza Trindade, G. Zoonotic Brazilian Vaccinia virus: From field to therapy. Antivir. Res. 2011, 92, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Stanford, M.M.; McFadden, G. The ‘supervirus’? Lessons from IL-4-expressing poxviruses. Trends Immunol. 2005, 6, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Castrucci, M.R.; Facchini, M.; Di Mario, G.; Garulli, B.; Sciaraffia, E.; Meola, M.; Fabiani, C.; De Marco, M.A.; Cordioli, P.; Siccardi, A.; et al. Modified vaccinia virus Ankara expressing the hemagglutinin of pandemic (H1N1) 2009 virus induces cross-protective immunity against Eurasian “avian-like” H1N1 swine viruses in mice. Influenza Other Respir. Viruses 2014, 8, 367–375. [Google Scholar] [CrossRef] [PubMed]
- García-Arriaza, J.; Cepeda, V.; Hallengärd, D.; Sorzano, C.Ó.; Kümmerer, B.M.; Liljeström, P.; Esteban, M. A Novel Poxvirus-Based Vaccine, MVA-CHIKV, Is Highly Immunogenic and Protects Mice against Chikungunya Infection. J. Virol. 2014, 88, 3527–3547. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.M.; Rahman, M.M.; McFadden, G. Oncolytic myxoma virus: The path to clinic. Vaccine 2013, 31, 4252–4258. [Google Scholar] [CrossRef] [PubMed]
- Moss, B. Poxvirus DNA Replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a010199. [Google Scholar] [CrossRef] [PubMed]
- Lefkowitz, E.J.; Wang, C.; Upton, C. Poxviruses: Past, present and future. Virus Res. 2006, 117, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Tulman, E.R.; Afonso, C.L.; Lu, Z.; Zsak, L.; Kutish, G.F.; Rock, D.L. The Genome of Canarypox Virus. J. Virol. 2004, 78, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Günther, T.; Haas, L.; Alawi, M.; Wohlsein, P.; Marks, J.; Grundhoff, A.; Becher, P.; Fischer, N. Recovery of the first full-length genome sequence of a parapoxvirus directly from a clinical sample. Sci. Rep. 2017, 7, 3734. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L.; Friedman, R. Poxvirus genome evolution by gene gain and loss. Mol. Phylogenet. Evol. 2005, 35, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Bratke, K.A.; McLysaght, A. Identification of multiple independent horizontal gene transfers into poxviruses using a comparative genomics approach. BMC Evol. Biol. 2008, 8, 67. [Google Scholar] [CrossRef] [PubMed]
- McLysaght, A.; Baldi, P.F.; Gaut, B.S. Extensive gene gain associated with adaptive evolution of poxviruses. Proc. Natl. Acad. Sci. USA 2003, 100, 15655–15660. [Google Scholar] [CrossRef] [PubMed]
- McFadden, G. Poxvirus tropism. Nat. Rev. Microbiol. 2005, 3, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Haller, S.L.; Peng, C.; McFadden, G.; Rothenburg, S. Poxviruses and the evolution of host range and virulence. Infect. Genet. Evol. 2014, 21, 15–40. [Google Scholar] [CrossRef] [PubMed]
- Bratke, K.A.; McLysaght, A.; Rothenburg, S. A survey of host range genes in poxvirus genomes. Infect. Genet. Evol. 2013, 14, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Werden, S.J.; Rahman, M.M.; McFadden, G. Poxvirus Host Range Genes Chapter 3. Adv. Virus Res. 2008, 71, 135–171. [Google Scholar] [PubMed]
- Chen, W.; Drillien, R.; Spehner, D.; Buller, R.M. In vitro and in vivo study of the ectromelia virus homolog of the vaccinia virus K1L host range gene. Virology 1993, 196, 682–693. [Google Scholar] [CrossRef] [PubMed]
- Stern, R.J.; Thompson, J.P.; Moyer, R.W. Attenuation of B5R mutants of rabbitpox virus in vivo is related to impaired growth and not an enhanced host inflammatory response. Virology 1997, 233, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wennier, S.; Moussatche, N.; Reinhard, M.; Condit, R.; McFadden, G. Myxoma Virus M064 Is a Novel Member of the Poxvirus C7L Superfamily of Host Range Factors That Controls the Kinetics of Myxomatosis in European Rabbits. J. Virol. 2012, 86, 5371–5375. [Google Scholar] [CrossRef] [PubMed]
- Damon, I.K. Poxviruses. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott, Williams and Wilkins: Philadelphia, PA, USA, 2014; Volume 2, p. 2160. [Google Scholar]
- Trindade, G.S.; Emerson, G.L.; Sammons, S.; Frace, M.; Govil, D.; Mota, B.E.F.; Abrahão, J.S.; de Assis, F.L.; Olsen-Rasmussen, M.; Goldsmith, C.S.; et al. Serro 2 Virus Highlights the Fundamental Genomic and Biological Features of a Natural Vaccinia Virus Infecting Humans. Viruses 2016, 8, 328. [Google Scholar] [CrossRef] [PubMed]
- Alzhanova, D.; Früh, K. Modulation of the host immune response by cowpox virus. Microbes Infect. 2010, 12, 900–909. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Anstey, A.V.; Bugert, J.J. Molluscum contagiosum virus infection. Lancet Infect. Dis. 2013, 10, 877–888. [Google Scholar] [CrossRef]
- Fenner, F. Adventures with poxviruses of vertebrates. FEMS Microbiol. Rev. 2000, 24, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Afonso, C.L.; Tulman, E.R.; Lu, Z.; Oma, E.; Kutish, G.F.; Rock, D.L. The Genome of Melanoplus sanguinipes Entomopoxvirus. J. Virol. 1999, 73, 533–552. [Google Scholar] [PubMed]
- Thézé, J.; Takatsuka, J.; Li, Z. New Insights into the Evolution of Entomopoxvirinae from the Complete Genome Sequences of Four Entomopoxviruses Infecting Adoxophyes honmai, Choristoneura biennis, Choristoneura rosaceana, and Mythimna separata. J. Virol. 2013, 87, 7992–8003. [Google Scholar] [CrossRef] [PubMed]
- Thézé, J.; Takatsuka, J.; Nakai, M.; Arif, B.; Herniou, E.A. Gene Acquisition Convergence between Entomopoxviruses and Baculoviruses. Viruses 2015, 7, 1960–1974. [Google Scholar] [CrossRef] [PubMed]
- Baker, K.S.; Leggett, R.M.; Bexfield, N.H.; Alston, M.; Daly, G.; Todd, S.; Tachedjian, M.; Holmes, C.E.; Crameri, S.; Wang, L.F. Metagenomic study of the viruses of African straw-coloured fruit bats: Detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virology 2013, 441, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Abrahão, J.S.; Campos, R.K.; De Souza Trindade, G.; Da Fonseca, F.G.; Ferreira, P.C.P.; Kroon, E.G. Outbreak of severe zoonotic vaccinia virus infection, Southeastern Brazil. Emerg. Infect. Dis. 2015, 21, 695–698. [Google Scholar] [CrossRef] [PubMed]
- Assis, F.L.; Borges, I.A.; Mesquita, V.S.; Ferreira, P.C.; Trindade, G.S.; Kroon, E.G.; Abrahão, J.S. Vaccinia virus in household environment during bovine vaccinia outbreak, Brazil. Emerg. Infect. Dis. 2013, 19, 2045–2047. [Google Scholar] [CrossRef] [PubMed]
- Nolen, L.D.; Osadebe, L.; Katomba, J.; Likofata, J.; Mukadi, D.; Monroe, B.; Doty, J.; Hughes, C.M.; Kabamba, J.; Malekani, J. Extended human-to-human transmission during a monkeypox outbreak in the Democratic Republic of the Congo. Emerg. Infect. Dis. 2016, 22, 1014–1021. [Google Scholar] [CrossRef] [PubMed]
- Campe, H.; Zimmermann, P.; Glos, K.; Bayer, M.; Bergemann, H.; Dreweck, C.; Graf, P.; Weber, B.K.; Meyer, H.; Büttner, M. Cowpox virus transmission from pet rats to humans, Germany. Emerg. Infect. Dis. 2009, 15, 777–780. [Google Scholar] [CrossRef] [PubMed]
- Shchelkunov, S.N. An Increasing Danger of Zoonotic Orthopoxvirus Infections. PLoS Pathog. 2013, 9, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Mayr, A.; Mahnel, H. Characterization of a fowlpox virus isolated from a rhinoceros. Arch. Gesamte Virusforsch. 1970, 31, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Droit, L.; Tesh, R.B.; Popov, V.L.; Little, N.S.; Upton, C.; Virgin, H.W.; Wang, D. The Genome of Yoka Poxvirus. J. Virol. 2011, 85, 10230–10238. [Google Scholar] [CrossRef] [PubMed]
- Abrahão, J.S.; Guedes, M.I.M.; Trindade, G.S.; Fonseca, F.G.; Campos, R.K.; Mota, B.F.; Lobato, Z.I.P.; Silva-Fernandes, A.T.; Rodrigues, G.O.L.; Lima, L.S. One more piece in the VACV ecological puzzle: Could peridomestic rodents be the link between wildlife and bovine vaccinia outbreaks in Brazil? PLoS ONE 2009, 4, 7428. [Google Scholar] [CrossRef] [PubMed]
- Hutson, C.L.; Nakazawa, Y.J.; Self, J.; Olson, V.A.; Regnery, R.L.; Braden, Z.; Weiss, S.; Malekani, J.; Jackson, E.; Tate, M. Laboratory Investigations of African Pouched Rats (Cricetomys gambianus) as a Potential Reservoir Host Species for Monkeypox Virus. PLoS Negl. Trop. Dis. 2015, 9, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Villarreal, L.P.; Defilippis, V.R.; Gottlieb, K.A. Acute and persistent viral life strategies and their relationship to emerging diseases. Virology 2000, 272, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Mauldin, M.; Antwerpen, M.; Emerson, G.; Li, Y.; Zoeller, G.; Carroll, D.; Meyer, H. Cowpox virus: What’s in a Name? Viruses 2017, 9, 101. [Google Scholar] [CrossRef] [PubMed]
- Kroon, E.G.; Abrahão, J.S.; de Souza Trindade, G.; Oliveira, G.P.; Luiz, A.P.M.F.; Costa, G.B.; Lima, M.T.; Calixto, R.S.; Oliveira, D.B.; Drumond, B.P. Natural Vaccinia virus infection: Diagnosis, isolation, and characterization. Curr. Protoc. Microbiol. 2016, 42, 14A.5.1–14A.5.43. [Google Scholar]
- Ninove, L.; Domart, Y.; Vervel, C.; Voinot, C.; Salez, N.; Raoult, D.; Meyer, H.; Capek, I.; Zandotti, C.; Charrel, R.N. Cowpox Virus Transmission from Pet Rats to Humans, France. Emerg. Infect. Dis. 2009, 15, 781–784. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.G.; Carroll, D.S.; Olson, V.A.; Hughes, C.; Galley, J.; Likos, A.; Montgomery, J.; Suu-Ire, R.; Kwasi, M.O.; Root, J.J. A silent enzootic of an orthopoxvirus in Ghana, West Africa: Evidence for multi-species involvement in the absence of widespread human disease. Am. J. Trop. Med. Hyg. 2010, 82, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Hutson, C.L.; Lee, K.N.; Abel, J.; Carroll, D.S.; Montgomery, J.M.; Olson, V.A.; Li, Y.; Davidson, W.; Hughes, C.; Dillon, M.; et al. Monkeypox zoonotic associations: Insights from laboratory evaluation of animals associated with the multi-state US outbreak. Am. J. Trop. Med. Hyg. 2007, 76, 757–768. [Google Scholar] [PubMed]
- Bowman, K.F.; Barbery, R.T.; Swango, L.J.; Schnurrenberger, P.R. Cutaneous form of bovine papular stommatitis in man. JAMA 1981, 246, 2813–2818. [Google Scholar] [CrossRef] [PubMed]
- Abrahão, J.S.; Silva-Fernandes, A.T.; Assis, F.L.; Guedes, M.I.; Drumond, B.P.; Leite, J.A.; Coelho, L.F.; Turrini, F.; Fonseca, F.G.; Lobato, Z.I. Human vaccinia virus and pseudocowpox virus co-infection: Clinical description and phylogenetic characterization. J. Clin. Virol. 2010, 48, 69–72. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.K.; Barankin, B.; Hon, K.L. Molluscum contagiosum: An update. Recent Pat. Inflamm. Allergy Drug Discov. 2017, 10, 2174. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, V.; Venkatesan, G.; Bhanuprakash, V.; Singh, R.K. Camelpox, an emerging orthopox viral disease. Indian J. Virol. 2013, 3, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, G.; Costa, G.; Assis, F.; Franco-Luiz, A.P.; Trindade, G.; Kroon, E.; Turrini, F.; Abrahão, J. Parapoxvirus. In Molecular Detection of Animal Viral Pathogens; Liu, D., Ed.; CRC Press: Boca Raton, FL, USA, 2016; pp. 881–890. [Google Scholar]
- Mendez-Rios, J.D.; Martens, C.A.; Bruno, D.P.; Porcella, S.F.; Zheng, Z.M.; Moss, B. Genome Sequence of Erythromelalgia-Related Poxvirus Identifies it as an Ectromelia Virus Strain. PLoS ONE 2012, 7, 34604. [Google Scholar] [CrossRef] [PubMed]
- Sigal, L.J. The pathogenesis and immunobiology of mousepox. Adv. Immunol. 2016, 129, 251–276. [Google Scholar] [PubMed]
- Franco-Luiz, A.P.; Oliveira, D.B.; Pereira, A.F.; Gasparini, M.C.; Bonjardim, C.A.; Ferreira, P.C.; Trindade, G.S.; Puentes, R.; Furtado, A.; Abrahão, J.S.; et al. Detection of Vaccinia Virus in Dairy Cattle Serum Samples from 2009, Uruguay. Emerg. Infect. Dis. 2016, 22, 2174–2177. [Google Scholar] [CrossRef] [PubMed]
- Friederichs, S.; Krebs, S.; Blum, H.; Lang, H.; Büttner, M. Parapoxvirus (PPV) of red deer reveals subclinical infection and confirms a unique species. J. Gen. Virol. 2015, 96, 1446–1462. [Google Scholar] [CrossRef] [PubMed]
- Fenner, F. The biological characters of several strains of vaccinia, cowpox, and rabbitpox viruses. Virology 1958, 5, 502–529. [Google Scholar] [CrossRef]
- Gemmell, A.; Cairns, J. Linkage in the genome of an animal virus. Virology 1959, 8, 381–383. [Google Scholar] [CrossRef]
- Sambrook, J.F.; Padgett, B.L.; Tomkins, J.K. Conditional lethal mutants of rabbitpox virus. I. Isolation of host cell-dependent and temperature-dependent mutants. Virology 1966, 28, 592–599. [Google Scholar] [CrossRef]
- Fenner, F.; Sambrook, J.F. Conditional lethal mutants of rabbitpox virus. II. Mutants (p) that fail to multiply in PK-2a cells. Virology 1966, 28, 600–609. [Google Scholar] [CrossRef]
- Ali, A.N.; Turner, P.C.; Brooks, M.A.; Moyer, R.W. The SPI-1 gene of rabbitpox virus determines host range and is required for hemorrhagic pock formation. Virology 1994, 202, 305–314. [Google Scholar] [CrossRef] [PubMed]
- McClain, M.E.; Greenland, R.M. Recombination between rabbitpox virus mutants in permissive and nonpermissive cells. Virology 1965, 25, 516–522. [Google Scholar] [CrossRef]
- Beattie, E.; Paoletti, E.; Tartaglia, J. Distinct patterns of IFN sensitivity observed in cells infected with vaccinia K3L- and E3L-mutant viruses. Virology 1965, 210, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.W.; Shun Chang, C.; Wang, G.; Werden, S.J.; Shao, Z.; Barrett, C.; Gao, X.; Belsito, T.A.; Villenevue, D.; McFadden, G. Myxoma virus M063R is a host range gene essential for virus replication in rabbit cells. Virology 2007, 361, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Legrand, F.A.; Verardi, P.H.; Jones, L.A.; Chan, K.S.; Peng, Y.; Yilma, T.D. Induction of potent humoral and cell-mediated immune responses by attenuated vaccinia virus vectors with deleted serpin genes. J. Virol. 2004, 78, 2770–2779. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.W.; Jacobs, B.L. Identification of a conserved motif that is necessary for binding of the vaccinia virus E3L gene products to double-stranded RNA. Virology 1993, 194, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.W.; Watson, J.C.; Jacobs, B.L. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. USA 1992, 89, 4825–4829. [Google Scholar] [CrossRef] [PubMed]
- Rivas, C.; Gil, J.; Melkova, Z.; Esteban, M.; Diaz-Guerra, M. Vaccinia virus E3Lprotein is an inhibitor of the interferon (i.f.n.)-induced 2–5A synthetase enzyme. Virology 1998, 243, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Langland, J.O.; Kash, J.C.; Carter, V.; Thomas, M.J.; Katze, M.G.; Jacobs, B.L. Suppression of proinflammatory signal transduction and gene expression by the dual nucleic acid binding domains of the vaccinia virus E3L proteins. J. Virol. 2006, 80, 10083–10095. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.V.; Elroy-Stein, O.; Jagus, R.; Moss, B.; Kaufman, R.J. The vaccinia virus K3L gene product potentiates translation by inhibiting double-stranded-RNA-activated protein kinase and phosphorylation of the alpha subunit of eukaryotic initiation factor 2. J. Virol. 1992, 66, 1943–1950. [Google Scholar] [PubMed]
- Davies, M.V.; Chang, H.W.; Jacobs, B.L.; Kaufman, R.J. The E3L and K3L vacciniavirus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J. Virol. 1993, 67, 1688–1692. [Google Scholar] [PubMed]
- Craig, A.W.; Cosentino, G.P.; Donze, O.; Sonenberg, N. The kinase insert domain of interferon-induced protein kinase PKR is required for activity but not for interactionwith the pseudosubstrate K3L. J. Biol. Chem. 1996, 271, 24526–24533. [Google Scholar] [CrossRef] [PubMed]
- Kawagishi-Kobayashi, M.; Silverman, J.B.; Ung, T.L.; Dever, T.E. Regulation of the protein kinase PKR by the vaccinia virus pseudosubstrate inhibitor K3L is dependenton residues conserved between the K3L protein and the PKR substrate eIF2alpha. Mol. Cell. Biol. 1997, 17, 4146–4158. [Google Scholar] [CrossRef] [PubMed]
- Langland, J.O.; Jacobs, B.L. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology 2002, 299, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Silverman, G.A.; Bird, P.I.; Carrell, R.W.; Church, F.C.; Coughlin, P.B.; Gettins, P.G.; Irving, J.A.; Lomas, D.A.; Luke, C.J.; Moyer, R.W. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 2001, 276, 33293–33296. [Google Scholar] [CrossRef] [PubMed]
- Herrera, E.; Lorenzo, M.M.; Blasco, R.; Isaacs, S.N. Functional analysis of vaccinia virus B5R protein: Essential role in virus envelopment is independent of a large portion of the extracellular domain. J. Virol. 1998, 72, 294–302. [Google Scholar] [PubMed]
- Rodger, G.; Smith, G.L. Replacing the SCR domains of vaccinia virus protein B5R with EGFP causes a reduction in plaque size and actin tail formation but enveloped virions are still transported to the cell surface. J. Gen. Virol. 2002, 83, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Spehner, D.; Gillard, S.; Drillien, R.; Kirn, A. A cowpox virus gene required for multiplication in Chinese hamster ovary cells. J. Virol. 1988, 62, 1297–1304. [Google Scholar] [PubMed]
- Chang, S.J.; Hsiao, J.C.; Sonnberg, S.; Chiang, C.T.; Yang, M.H.; Tzou, D.L.; Mercer, A.A.; Chang, W. Poxviral host range protein CP77 contains a F-box-like domain that is necessary to suppress NF-{kappa}B activation by TNF-{alpha} but is independent of its host range function. J. Virol. 2009, 83, 4140–4152. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Chao, J.; Xiang, Y. Identification from diverse mammalian poxviruses of host-range regulatory genes functioning equivalently to vaccinia virus C7L. Virology 2008, 372, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Davis, T.; Anderson, D.; Solam, L.; Beckmann, M.P.; Jerzy, R.; Dower, S.K.; Cosman, D.; Goodwin, R.G. A receptor for tumor necrosis factor defines an unusual family of cellular and viral proteins. Science 1990, 248, 1019–1023. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Davis, T.; Wignall, J.M.; Din, W.S.; Farrah, T.; Upton, C.; McFadden, G.; Goodwin, R.G. T2 open reading frame from the Shope fibroma virus encodes a soluble form of the TNF receptor. Biochem. Biophys. Res. Commun. 1991, 176, 335–342. [Google Scholar] [CrossRef]
- Alejo, A.; Ruiz-Arguello, M.B.; Ho, Y.; Smith, V.P.; Saraiva, M.; Alcami, A. A chemokine-binding domain in the tumor necrosis factor receptor from variola (smallpox) virus. Proc. Natl. Acad. Sci. USA 2006, 103, 5995–6000. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.E.; Corbett, K.D.; Berger, J.M.; McFadden, G.; Handel, T.M. Structure of M11L: A myxoma virus structural homolog of the apoptosis inhibitor, Bcl-2. Protein Sci. 2007, 16, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Johnston, J.B.; Barrett, J.W.; Nazarian, S.H.; Goodwin, M.; Ricciuto, D.; Wang, G.; McFadden, G. A poxvirus-encoded pyrin domain protein interacts with ASC-1 to inhibit host inflammatory and apoptotic responses to infection. Immunity 2005, 23, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Senkevich, T.G.; Koonin, E.V.; Bugert, J.J.; Darai, G.; Moss, B. The genome of molluscum contagiosum virus: Analysis and comparison with other poxviruses. Virology 1997, 233, 19–42. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L.; Irausquin, S.; Friedman, R. The evolutionary biology of poxviruses. Infect. Genet. Evol. 2010, 10, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Aravind, L.; Koonin, E.V. Common Origin of Four Diverse Families of Large Eukaryotic DNA Viruses. J. Virol. 2001, 75, 11720–11734. [Google Scholar] [CrossRef] [PubMed]
- Boyer, M.; Gimenez, G.; Suzan-Monti, M.; Raoult, D. Classification and determination of possible origins of ORFans through analysis of nucleocytoplasmic large DNA viruses. Intervirology 2010, 53, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L. Origin and evolution of viral interleukin-10 and other DNA virus genes with vertebrate homologues. J. Mol. Evol. 2002, 54, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Smithson, C.; Meyer, H.; Gigante, C.M.; Gao, J.; Zhao, H.; Batra, D.; Damon, I.; Upton, C.; Li, Y. Two novel poxviruses with unusual genome rearrangements: NY_014 and Murmansk. Virus Genes 2017. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.K.; Young, V.L.; Kleffmann, T.; Ward, V.K. Genomic and proteomic analysis of invertebrate iridovirus type 9. J. Virol. 2011, 85, 7900–7911. [Google Scholar] [CrossRef] [PubMed]
- Legendre, M.; Santini, S.; Rico, A.; Abergel, C.; Claverie, J.M. Breaking the 1000-gene barrier for Mimivirus using ultra-deep genome and transcriptome sequencing. Virol. J. 2011, 8, 99. [Google Scholar] [CrossRef] [PubMed]
- Odom, M.R.; Hendrickson, R.C.; Lefkowitz, E.J. Poxvirus protein evolution: Family-wide assessment of possible horizontal gene transfer events. Virus Res. 2009, 144, 233–249. [Google Scholar] [CrossRef] [PubMed]
- Gjessing, M.C.; Yutin, N.; Tengs, T.; Senkevich, T.; Koonin, E.; Rønning, H.P.; Alarcon, M.; Ylving, S.; Lie, K.I.; Saure, B. Salmon Gill Poxvirus, the Deepest Representative of the Chordopoxvirinae. J. Virol. 2015, 89, 9348–9367. [Google Scholar] [CrossRef] [PubMed]
- O’Dea, M.; Tu, S.-L.; Pang, S.; De Ridder, T.; Jackson, B.; Upton, C. Genomic characterization of a novel poxvirus from a flying fox: Evidence for a new genus? J. Gen. Virol. 2016, 97, 2363–2375. [Google Scholar] [CrossRef] [PubMed]
- Tu, S.L.; Nakazawa, Y.; Gao, J.; Wilkins, K.; Gallardo-Romero, N.; Li, Y.; Emerson, G.L.; Carroll, D.S.; Upton, C. Characterization of Eptesipoxvirus, a novel poxvirus from a microchiropteran bat. Virus Genes 2017. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Senkevich, T.G.; Dolja, V.V. The ancient Virus World and evolution of cells. Biol. Direct 2006, 1, 29. [Google Scholar] [CrossRef] [PubMed]
- Barry, M.; McFadden, G. Virus encoded cytokines and cytokine receptors. Parasitology 1997, 115, 89–100. [Google Scholar] [CrossRef]
- Lawrence, J.G.; Ochman, H. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. USA 1998, 95, 9413–9417. [Google Scholar] [CrossRef] [PubMed]
- Ochman, H.; Lawrence, J.G.; Groisman, E.A. Lateral gene transfer and the nature of bacterial innovation. Nature 2000, 405, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Upton, C.; McFadden, G. DNA sequence homology between the terminal inverted repeats of Shope fibroma virus and an endogenous cellular plasmid species. Mol. Cell. Biol. 1986, 6, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Liszewski, M.K.; Leung, M.K.; Hauhart, R.; Fang, C.J.; Bertram, P.; Atkinson, J.P. Smallpox inhibitor of complement enzymes (SPICE): Dissecting functional sites and abrogating activity. J. Immunol. 2009, 183, 3150–3159. [Google Scholar] [CrossRef] [PubMed]
- Ojha, H.; Panwar, H.S.; Gorham, R.D.; Morikis, D.; Sahu, A. Viral regulators of complement activation: Structure, function and evolution. Mol. Immunol. 2014, 61, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Kotwal, G.J.; Moss, B. Analysis of a large cluster of nonessential genes deleted from a vaccinia virus terminal transposition mutant. Virology 1988, 167, 524–537. [Google Scholar] [CrossRef]
- Kotwal, G.J.; Isaacs, S.N.; McKenzie, R.; Frank, M.M.; Moss, B. Inhibition of the complement cascade by the major secretory protein of vaccinia virus. Science 1990, 250, 827–830. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, G.P.; Rodrigues, R.A.L.; Lima, M.T.; Drumond, B.P.; Abrahão, J.S. Poxvirus Host Range Genes and Virus–Host Spectrum: A Critical Review. Viruses 2017, 9, 331. https://doi.org/10.3390/v9110331
Oliveira GP, Rodrigues RAL, Lima MT, Drumond BP, Abrahão JS. Poxvirus Host Range Genes and Virus–Host Spectrum: A Critical Review. Viruses. 2017; 9(11):331. https://doi.org/10.3390/v9110331
Chicago/Turabian StyleOliveira, Graziele Pereira, Rodrigo Araújo Lima Rodrigues, Maurício Teixeira Lima, Betânia Paiva Drumond, and Jônatas Santos Abrahão. 2017. "Poxvirus Host Range Genes and Virus–Host Spectrum: A Critical Review" Viruses 9, no. 11: 331. https://doi.org/10.3390/v9110331




