Deletion of A44L, A46R and C12L Vaccinia Virus Genes from the MVA Genome Improved the Vector Immunogenicity by Modifying the Innate Immune Response Generating Enhanced and Optimized Specific T-Cell Responses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Construction of Deleted MVAs: MVA∆A44L-A46R and MVA∆C12L/∆A44L-A46R
RT-PCRs
2.3. Viral Immunization Stocks
In Vitro Characterization of MVA∆A44L-A46R and MVA∆C12L/∆A44L-A46R Viruses
2.4. THP-1 Infection and Cytokine Production Evaluation
2.5. 3β-HSD Activity
2.6. IL-18 Binding-Protein Bioassay
2.7. Immunization Protocols, Sample Collection, and Processing
2.8. Analysis of Specific T-Cell Immune Responses
2.8.1. Peptides
2.8.2. Murine IFN-γ and IL-2 ELISPOT Assays
2.8.3. Intracellular Cytokine Staining (ICS) and Proliferation Assays
2.8.4. T-Cell Specific Cytokine Production
2.8.5. In Vitro Cytotoxicity Assays
2.9. Innate Immune Response Analyses
2.10. Data Analysis
3. Results
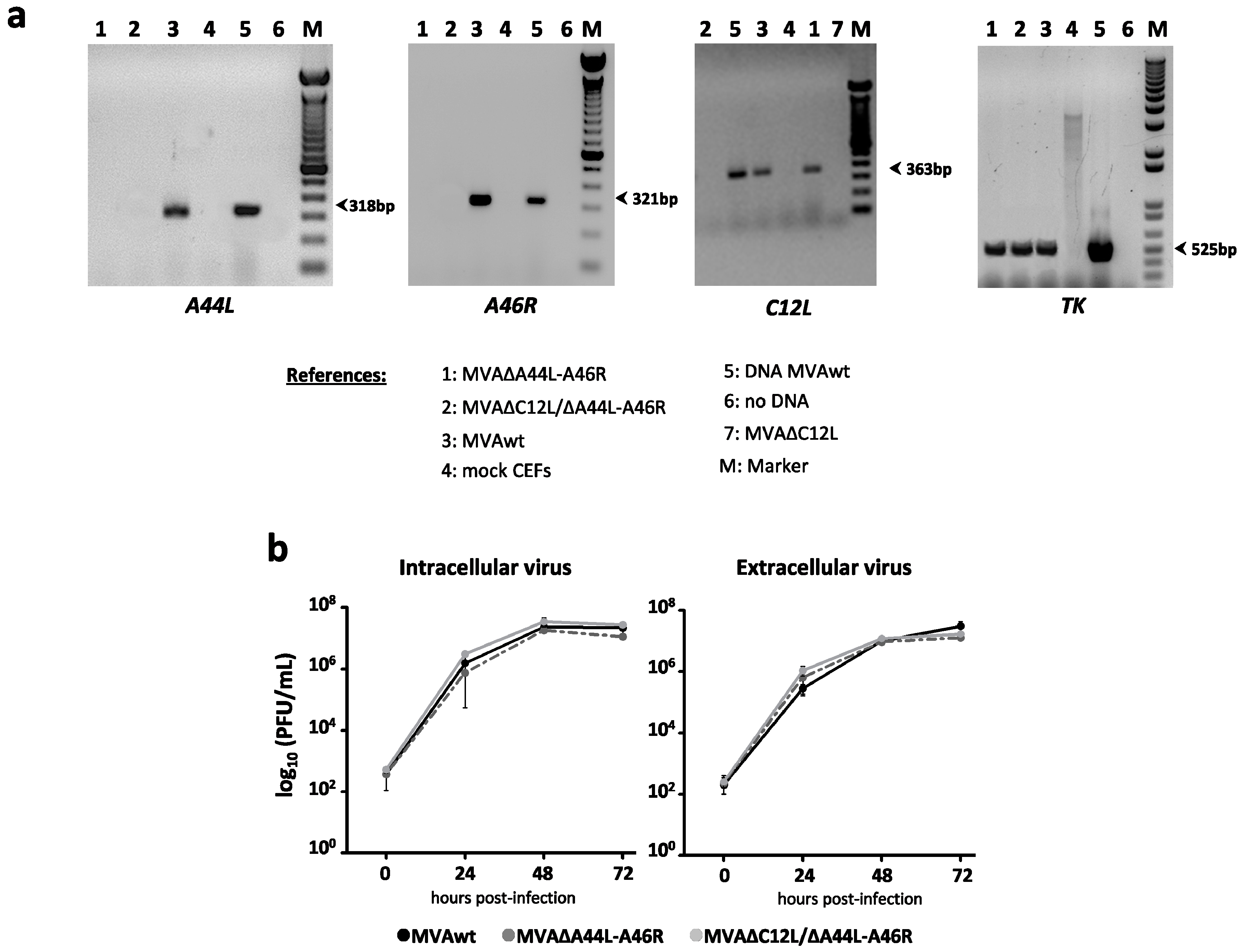
3.1. In Vitro Characterization of MVA Vectors after Deleting A44L, A46R, and C12L Genes
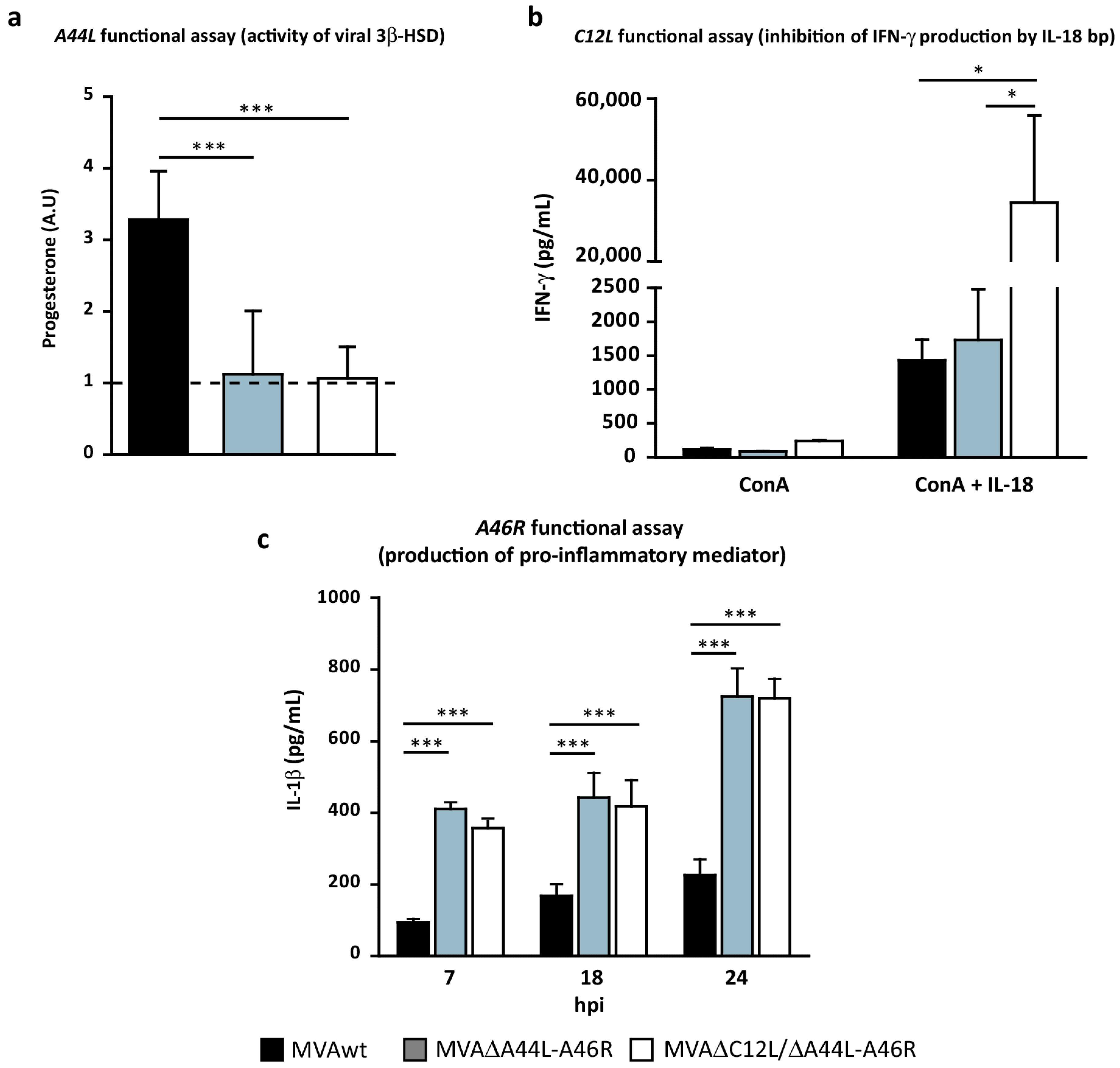
3.2. In Vitro Analysis of the Biological Effects of Deleted Viral Genes
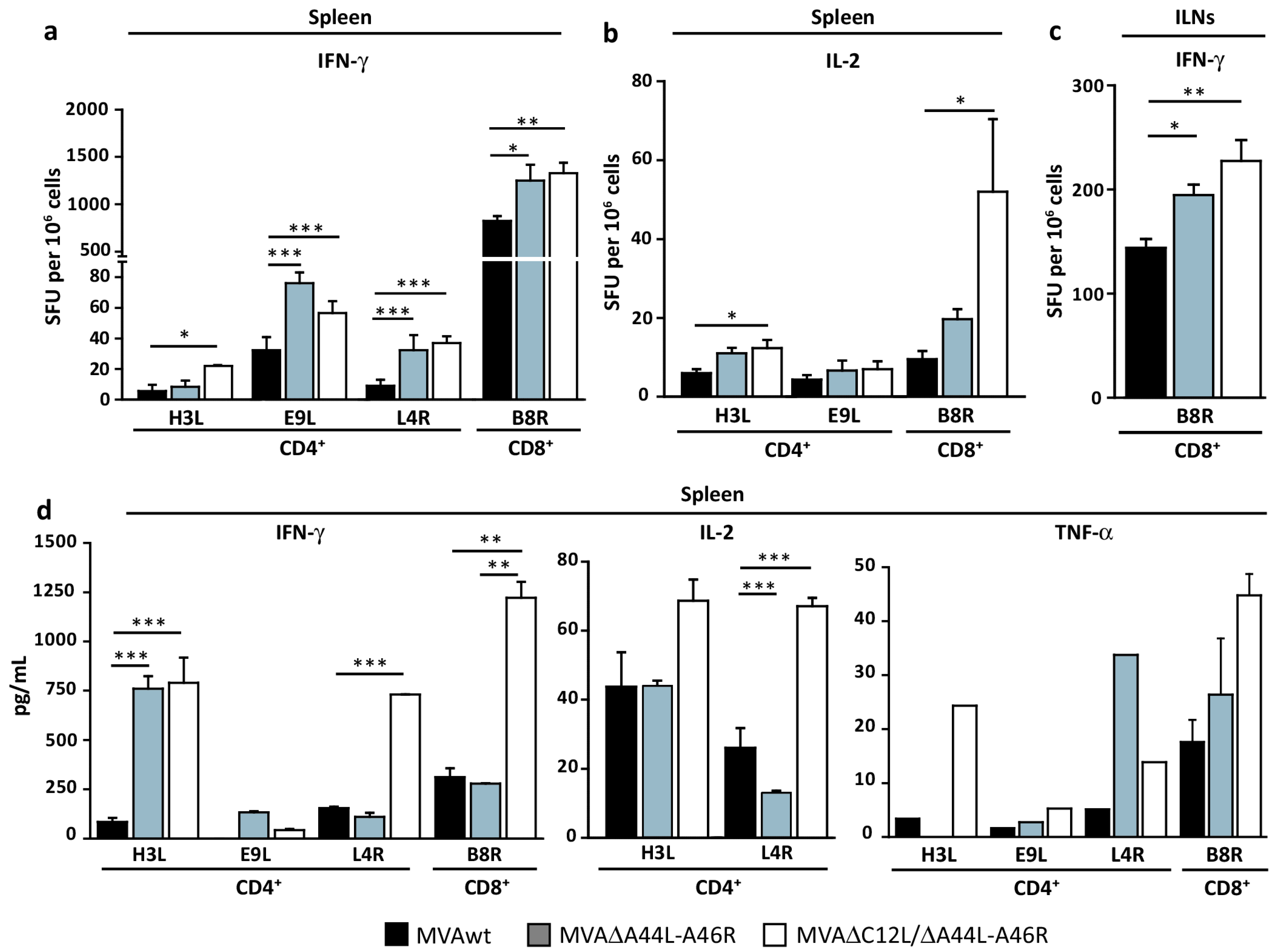
3.3. Modulation of Specific T-Cell Responses by MVA∆A44L-A46R and MVA∆C12L/∆A44L-A46R Vectors
3.4. At Later Times Post-Immunization Deleted MVAs Still Induced Higher Specific T-Cellular Responses
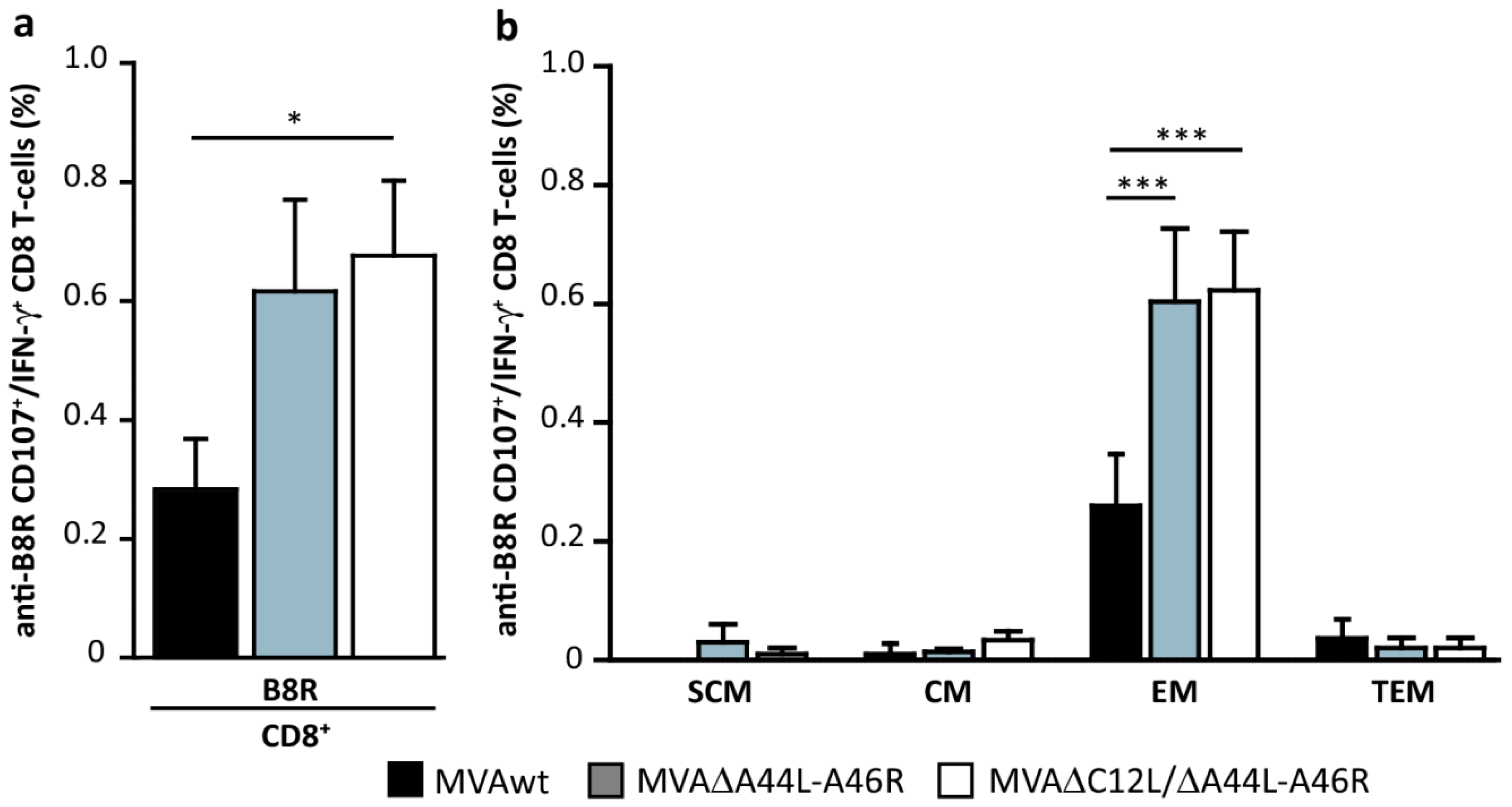
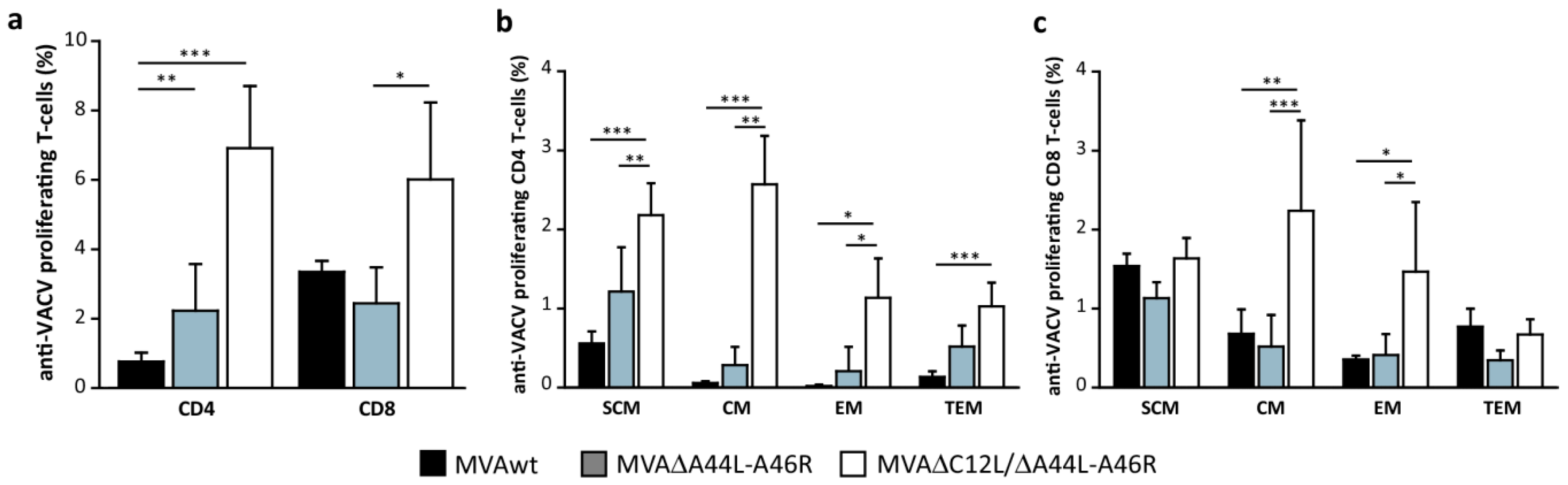
3.5. MVA∆A44L-A46R and MVA∆C12L/∆A44L-A46R Vectors Improved the Quality of Memory T-Cell Responses
3.6. Pattern of Cytokines Induced during Innate Immune Response by MVA∆C12L/∆A44L-A46R
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CEF | chicken embryo fibroblasts |
| ConA | Concanavalin A |
| CVA | Chorioallantois vaccinia virus Ankara |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| DMSO | dimethylsulfoxide |
| dpi | days post-infection |
| FBS | fetal bovineserum |
| GUS | β-glucuronidase enzyme |
| hpi | hours post-infection |
| ICS | Intracellular cytokine staining |
| MVA | Modified Vaccinia Ankara Virus |
| moi | multiplicity of infection |
| NYVAC | New York Vaccinia virus |
| ORF | open reading frames |
| PFU | plaque former units |
| PMA | phorbol-12-myristate-13-acetate |
| SD | Standard deviation |
| SFU | Spot-forming unit |
| SPF | specific pathogen free |
| TCM | T-cell Central Memory |
| TEM | T-cell Effector Memory |
| TSCM | T-cell Stem Cell Memory |
| TTEM | T-cell Terminally Effector Memory |
| TIR | Toll/interleukin-1 receptor |
| TLR | Toll Like Receptor |
| VACV | Vaccinia virus |
| WR | Western Reserve strain |
| 3β-HSD | 3β-hydroxysteroid-dehydrogenase/∆5-∆4 isomerase |
References
- Drexler, I.; Heller, K.; Wahren, B.; Erfle, V.; Sutter, G. Highly attenuated modified vaccinia virus Ankara replicates in baby hamster kidney cells, a potential host for virus propagation, but not in various human transformed and primary cells. J. Gener. Virol. 1998, 79, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Carroll, M.W.; Moss, B. Host range and cytopathogenicity of the highly attenuated MVA strain of vaccinia virus: Propagation and generation of recombinant viruses in a nonhuman mammalian cell line. Virology 1997, 238, 198–211. [Google Scholar] [CrossRef] [PubMed]
- McCurdy, L.H.; Larkin, B.D.; Martin, J.E.; Graham, B.S. Modified vaccinia Ankara: Potential as an alternative smallpox vaccine. Clin. Infect. Dis. 2004, 38, 1749–1753. [Google Scholar] [CrossRef] [PubMed]
- Antoine, G.; Scheiflinger, F.; Dorner, F.; Falkner, F.G. The complete genomic sequence of the modified vaccinia Ankara strain: Comparison with other orthopoxviruses. Virology 1998, 244, 365–396. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, T.J.; Alcami, A.; Andrea, P.; Smith, G.L. Modified vaccinia virus Ankara undergoes limited replication in human cells and lacks several immunomodulatory proteins: Implications for use as a human vaccine. J. Gener. Virol. 1998, 79, 1159–1167. [Google Scholar] [CrossRef] [PubMed]
- Dobson, B.M.; Tscharke, D.C. Truncation of gene F5L partially masks rescue of vaccinia virus strain MVA growth on mammalian cells by restricting plaque size. J. Gener. Virol. 2014, 95, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Gomez, C.E.; Perdiguero, B.; Garcia-Arriaza, J.; Esteban, M. Clinical applications of attenuated MVA poxvirus strain. Exp. Rev. Vaccines 2013, 12, 1395–1416. [Google Scholar] [CrossRef] [PubMed]
- Acres, B.; Bonnefoy, J.Y. Clinical development of MVA-based therapeutic cancer vaccines. Exp. Rev. Vaccines 2008, 7, 889–893. [Google Scholar] [CrossRef] [PubMed]
- Sutter, G.; Staib, C. Vaccinia vectors as candidate vaccines: The development of modified vaccinia virus Ankara for antigen delivery. Curr. Drug Targets Infect. Disord. 2003, 3, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S.C. Clinical development of modified vaccinia virus Ankara vaccines. Vaccine 2013, 31, 4241–4246. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.V.; Reyes-Sandoval, A.; O’Hara, G.; Ewer, K.; Lawrie, A.; Goodman, A.; Nicosia, A.; Folgori, A.; Colloca, S.; Cortese, R.; et al. Prime-boost vectored malaria vaccines: Progress and prospects. Hum. Vaccines 2010, 6, 78–83. [Google Scholar] [CrossRef]
- Bejon, P.; Ogada, E.; Mwangi, T.; Milligan, P.; Lang, T.; Fegan, G.; Gilbert, S.C.; Peshu, N.; Marsh, K.; Hill, A.V. Extended follow-up following a phase 2b randomized trial of the candidate malaria vaccines FP9 ME-TRAP and MVA ME-TRAP among children in Kenya. PLoS ONE 2007, 2, e707. [Google Scholar] [CrossRef] [PubMed]
- Hawkridge, T.; Scriba, T.J.; Gelderbloem, S.; Smit, E.; Tameris, M.; Moyo, S.; Lang, T.; Veldsman, A.; Hatherill, M.; Merwe, L.; et al. Safety and immunogenicity of a new tuberculosis vaccine, MVA85A, in healthy adults in South Africa. J. Infect. Dis. 2008, 198, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Nicol, M.P.; Grobler, L.A. MVA-85A, a novel candidate booster vaccine for the prevention of tuberculosis in children and adults. Curr. Opin. Mol. Ther. 2010, 12, 124–134. [Google Scholar] [PubMed]
- Kim, D.W.; Krishnamurthy, V.; Bines, S.D.; Kaufman, H.L. TroVax, a recombinant modified vaccinia Ankara virus encoding 5T4: Lessons learned and future development. Hum. Vaccines 2010, 6, 784–791. [Google Scholar] [CrossRef]
- Dangoor, A.; Lorigan, P.; Keilholz, U.; Schadendorf, D.; Harris, A.; Ottensmeier, C.; Smyth, J.; Hoffmann, K.; Anderson, R.; Cripps, M.; et al. Clinical and immunological responses in metastatic melanoma patients vaccinated with a high-dose poly-epitope vaccine. Cancer Immunol. Immunother. 2010, 59, 863–873. [Google Scholar] [CrossRef] [PubMed]
- Meisinger-Henschel, C.; Schmidt, M.; Lukassen, S.; Linke, B.; Krause, L.; Konietzny, S.; Goesmann, A.; Howley, P.; Chaplin, P.; Suter, M.; et al. Genomic sequence of chorioallantois vaccinia virus Ankara, the ancestor of modified vaccinia virus Ankara. J. Gener. Virol. 2007, 88, 3249–3259. [Google Scholar] [CrossRef] [PubMed]
- Staib, C.; Kisling, S.; Erfle, V.; Sutter, G. Inactivation of the viral interleukin 1β receptor improves CD8+ T-cell memory responses elicited upon immunization with modified vaccinia virus Ankara. J. Gener. Virol. 2005, 86, 1997–2006. [Google Scholar] [CrossRef] [PubMed]
- Clark, R.H.; Kenyon, J.C.; Bartlett, N.W.; Tscharke, D.C.; Smith, G.L. Deletion of gene A41L enhances vaccinia virus immunogenicity and vaccine efficacy. J. Gener. Virol. 2006, 87, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Falivene, J.; del Medico Zajac, M.P.; Pascutti, M.F.; Rodriguez, A.M.; Maeto, C.; Perdiguero, B.; Gomez, C.E.; Esteban, M.; Calamante, G.; Gherardi, M.M. Improving the MVA vaccine potential by deleting the viral gene coding for the IL-18 binding protein. PLoS ONE 2012, 7, e32220. [Google Scholar] [CrossRef] [PubMed]
- Perdiguero, B.; Gomez, C.E.; Najera, J.L.; Sorzano, C.O.; Delaloye, J.; Gonzalez-Sanz, R.; Jimenez, V.; Roger, T.; Calandra, T.; Pantaleo, G.; et al. Deletion of the viral anti-apoptotic gene F1L in the HIV/AIDS vaccine candidate MVA-C enhances immune responses against HIV-1 antigens. PLoS ONE 2012, 7, e48524. [Google Scholar] [CrossRef] [PubMed]
- Garber, D.A.; O’Mara, L.A.; Gangadhara, S.; McQuoid, M.; Zhang, X.; Zheng, R.; Gill, K.; Verma, M.; Yu, T.; Johnson, B.; et al. Deletion of specific immune-modulatory genes from modified vaccinia virus Ankara-based HIV vaccines engenders improved immunogenicity in Rhesus Macaques. J. Virol. 2012, 86, 12605–12615. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Arriaza, J.; Arnaez, P.; Gomez, C.E.; Sorzano, C.O.; Esteban, M. Improving adaptive and memory immune responses of an HIV/AIDS vaccine candidate MVA-B by deletion of vaccinia virus genes (C6L and K7R) blocking interferon signaling pathways. PLoS ONE 2013, 8, e66894. [Google Scholar]
- Garcia-Arriaza, J.; Gomez, C.E.; Sorzano, C.O.; Esteban, M. Deletion of the vaccinia virus N2L gene encoding an inhibitor of IRF3 improves the immunogenicity of modified vaccinia virus Ankara expressing HIV-1 antigens. J. Virol. 2014, 88, 3392–3410. [Google Scholar] [CrossRef] [PubMed]
- Bowie, A.; Kiss-Toth, E.; Symons, J.A.; Smith, G.L.; Dower, S.K.; O’Neill, L.A. A46R and A52R from vaccinia virus are antagonists of host IL-1 and toll-like receptor signaling. Proc. Natl. Acad. Sci. USA 2000, 97, 10162–10167. [Google Scholar] [CrossRef] [PubMed]
- Stack, J.; Haga, I.R.; Schroder, M.; Bartlett, N.W.; Maloney, G.; Reading, P.C.; Fitzgerald, K.A.; Smith, G.L.; Bowie, A.G. Vaccinia virus protein A46R targets multiple toll-like-interleukin-1 receptor adaptors and contributes to virulence. J. Exp. Med. 2005, 201, 1007–1018. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; May, M.J.; Kopp, E.B. Nf-κb and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 1998, 16, 225–260. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Takaoka, A.; Taniguchi, T. Type I interferon gene induction by the interferon regulatory factor family of transcription factors. Immunity 2006, 25, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Innate immunity to virus infection. Immunol. Rev. 2009, 227, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Reading, P.C.; Moore, J.B.; Smith, G.L. Steroid hormone synthesis by vaccinia virus suppresses the inflammatory response to infection. J. Exp. Med. 2003, 197, 1269–1278. [Google Scholar] [CrossRef] [PubMed]
- Boumpas, D.T.; Chrousos, G.P.; Wilder, R.L.; Cupps, T.R.; Balow, J.E. Glucocorticoid therapy for immune-mediated diseases: Basic and clinical correlates. Ann. Intern. Med. 1993, 119, 1198–1208. [Google Scholar] [CrossRef] [PubMed]
- Perdiguero, B.; Gomez, C.E.; di Pilato, M.; Sorzano, C.O.; Delaloye, J.; Roger, T.; Calandra, T.; Pantaleo, G.; Esteban, M. Deletion of the vaccinia virus gene A46R, encoding for an inhibitor of TLR signalling, is an effective approach to enhance the immunogenicity in mice of the HIV/AIDS vaccine candidate NYVAC-C. PLoS ONE 2013, 8, e74831. [Google Scholar]
- Alharbi, N.K.; Spencer, A.J.; Hill, A.V.; Gilbert, S.C. Deletion of fifteen open reading frames from modified vaccinia virus Ankara fails to improve immunogenicity. PLoS ONE 2015, 10, e0128626. [Google Scholar] [CrossRef] [PubMed]
- Cottingham, M.G.; Andersen, R.F.; Spencer, A.J.; Saurya, S.; Furze, J.; Hill, A.V.; Gilbert, S.C. Recombination-mediated genetic engineering of a bacterial artificial chromosome clone of modified vaccinia virus Ankara (MVA). PLoS ONE 2008, 3, e1638. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.B.; Smith, G.L. Steroid hormone synthesis by a vaccinia enzyme: A new type of virus virulence factor. EMBO J. 1992, 11, 3490. [Google Scholar] [PubMed]
- Sroller, V.; Kutinova, L.; Nemeckova, S.; Simonova, V.; Vonka, V. Effect of 3-β-hydroxysteroid dehydrogenase gene deletion on virulence and immunogenicity of different vaccinia viruses and their recombinants. Arch. Virol. 1998, 143, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- Tscharke, D.C.; Reading, P.C.; Smith, G.L. Dermal infection with vaccinia virus reveals roles for virus proteins not seen using other inoculation routes. J. Gener. Virol. 2002, 83, 1977–1986. [Google Scholar] [CrossRef] [PubMed]
- Almazan, F.; Tscharke, D.C.; Smith, G.L. The vaccinia virus superoxide dismutase-like protein (A45R) is a virion component that is nonessential for virus replication. J. Virol. 2001, 75, 7018–7029. [Google Scholar] [CrossRef] [PubMed]
- Mayr, A.; Stickl, H.; Muller, H.K.; Danner, K.; Singer, H. The smallpox vaccination strain MVA: Marker, genetic structure, experience gained with the parenteral vaccination and behavior in organisms with a debilitated defence mechanism (author’s transl). Zentralblatt fur Bakteriologie, Parasitenkunde, Infektionskrankheiten und Hygiene. Erste Abteilung Originale 1978, 167, 375–390. [Google Scholar]
- Earl, P.L.; Cooper, N.; Wyatt, L.S.; Moss, B.; Carroll, M.W. Preparation of cell cultures and vaccinia virus stocks. Curr. Protoc. Mol. Biol. 2001. [Google Scholar] [CrossRef]
- Ramirez, J.C.; Gherardi, M.M.; Esteban, M. Biology of attenuated modified vaccinia virus Ankara recombinant vector in mice: Virus fate and activation of B- and T-cell immune responses in comparison with the western reserve strain and advantages as a vaccine. J. Virol. 2000, 74, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Tscharke, D.C.; Karupiah, G.; Zhou, J.; Palmore, T.; Irvine, K.R.; Haeryfar, S.M.; Williams, S.; Sidney, J.; Sette, A.; Bennink, J.R.; et al. Identification of poxvirus CD8+ T cell determinants to enable rational design and characterization of smallpox vaccines. J. Exp. Med. 2005, 201, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.M.; Turk, G.; Pascutti, M.F.; Ferrer, F.; Najera, J.L.; Monaco, D.; Esteban, M.; Salomon, H.; Calamante, G.; Gherardi, M.M. Characterization of DNA and MVA vectors expressing Nef from HIV-1 CRF12_BF revealed high immune specificity with low cross-reactivity against subtype B. Virus Res. 2009, 146, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Betts, M.R.; Brenchley, J.M.; Price, D.A.; de Rosa, S.C.; Douek, D.C.; Roederer, M.; Koup, R.A. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J. Immunol. Methods 2003, 281, 65–78. [Google Scholar] [CrossRef]
- Maeto, C.; Rodriguez, A.M.; Holgado, M.P.; Falivene, J.; Gherardi, M.M. Novel mucosal DNA-MVA HIV vaccination in which DNA-IL-12 plus cholera toxin B subunit (CTB) cooperates to enhance cellular systemic and mucosal genital tract immunity. PLoS ONE 2014, 9, e107524. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.M.; Pascutti, M.F.; Maeto, C.; Falivene, J.; Holgado, M.P.; Turk, G.; Gherardi, M.M. IL-12 and GM-CSF in DNA/MVA immunizations against HIV-1 CRF12_BF Nef induced T-cell responses with an enhanced magnitude, breadth and quality. PLoS ONE 2012, 7, e37801. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, Y.; Watari, E.; Shimizu, M.; Takahashi, H. One-step simple assay to determine antigen-specific cytotoxic activities by single-color flow cytometry. Biomed. Res. 2011, 32, 159–166. [Google Scholar] [CrossRef] [PubMed]
- GraphPad Prism; Version 5.03; GraphPad Software Inc.: LA Jolla, CA, USA, 2009.
- Engelstad, M.; Howard, S.T.; Smith, G.L. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology 1992, 188, 801–810. [Google Scholar] [CrossRef]
- Roper, R.L.; Wolffe, E.J.; Weisberg, A.; Moss, B. The envelope protein encoded by the A33R gene is required for formation of actin-containing microvilli and efficient cell-to-cell spread of vaccinia virus. J. Virol. 1998, 72, 4192–4204. [Google Scholar] [PubMed]
- Abadie, V.; Bonduelle, O.; Duffy, D.; Parizot, C.; Verrier, B.; Combadiere, B. Original encounter with antigen determines antigen-presenting cell imprinting of the quality of the immune response in mice. PLoS ONE 2009, 4, e8159. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Joe, G.; Hexner, E.; Zhu, J.; Emerson, S.G. Host-reactive CD8+ memory stem cells in graft-versus-host disease. Nat. Med. 2005, 11, 1299–1305. [Google Scholar] [CrossRef] [PubMed]
- Gattinoni, L.; Zhong, X.S.; Palmer, D.C.; Ji, Y.; Hinrichs, C.S.; Yu, Z.; Wrzesinski, C.; Boni, A.; Cassard, L.; Garvin, L.M.; et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat. Med. 2009, 15, 808–813. [Google Scholar] [CrossRef] [PubMed]
- Seth, R.B.; Sun, L.; Chen, Z.J. Antiviral innate immunity pathways. Cell Res. 2006, 16, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Dai, P.; Wang, W.; Cao, H.; Avogadri, F.; Dai, L.; Drexler, I.; Joyce, J.A.; Li, X.D.; Chen, Z.; Merghoub, T.; et al. Modified vaccinia virus Ankara triggers type I IFN production in murine conventional dendritic cells via a cGAS/STING-mediated cytosolic DNA-sensing pathway. PLoS Pathog. 2014, 10, e1003989. [Google Scholar] [CrossRef] [PubMed]
- Kreijtz, J.H.; Gilbert, S.C.; Sutter, G. Poxvirus vectors. Vaccine 2013, 31, 4217–4219. [Google Scholar] [CrossRef] [PubMed]
- Delaloye, J.; Roger, T.; Steiner-Tardivel, Q.G.; le Roy, D.; Knaup Reymond, M.; Akira, S.; Petrilli, V.; Gomez, C.E.; Perdiguero, B.; Tschopp, J.; et al. Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS Pathog. 2009, 5, e1000480. [Google Scholar] [CrossRef] [PubMed]
- Appay, V.; Douek, D.C.; Price, D.A. CD8+ T cell efficacy in vaccination and disease. Nat. Med. 2008, 14, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Hickman, H.D.; Takeda, K.; Skon, C.N.; Murray, F.R.; Hensley, S.E.; Loomis, J.; Barber, G.N.; Bennink, J.R.; Yewdell, J.W. Direct priming of antiviral CD8+ T cells in the peripheral interfollicular region of lymph nodes. Nat. Immunol. 2008, 9, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Perdiguero, B.; Gomez, C.E.; Cepeda, V.; Sanchez-Sampedro, L.; Garcia-Arriaza, J.; Mejias-Perez, E.; Jimenez, V.; Sanchez, C.; Sorzano, C.O.; Oliveros, J.C.; et al. Virological and immunological characterization of novel NYVAC-based HIV/AIDS vaccine candidates expressing clade C trimeric soluble gp140 (ZM96) and Gag (ZM96)-Pol-Nef (CN54) as virus-like particles. J. Virol. 2015, 89, 970–988. [Google Scholar] [CrossRef] [PubMed]
- Delaloye, J.; Filali-Mouhim, A.; Cameron, M.J.; Haddad, E.K.; Harari, A.; Goulet, J.P.; Gomez, C.E.; Perdiguero, B.; Esteban, M.; Pantaleo, G.; et al. Interleukin-1- and type I interferon-dependent enhanced immunogenicity of an NYVAC-HIV-1 Env-Gag-Pol-Nef vaccine vector with dual deletions of type I and type II interferon-binding proteins. J. Virol. 2015, 89, 3819–3832. [Google Scholar] [CrossRef] [PubMed]
- Wolferstatter, M.; Schweneker, M.; Spath, M.; Lukassen, S.; Klingenberg, M.; Brinkmann, K.; Wielert, U.; Lauterbach, H.; Hochrein, H.; Chaplin, P.; et al. Recombinant modified vaccinia virus Ankara generating excess early double-stranded RNA transiently activates protein kinase R and triggers enhanced innate immune responses. J. Virol. 2014, 88, 14396–14411. [Google Scholar] [CrossRef] [PubMed]
- Watford, W.T.; Moriguchi, M.; Morinobu, A.; O’Shea, J.J. The biology of IL-12: Coordinating innate and adaptive immune responses. Cytokine Growth Factor Rev. 2003, 14, 361–368. [Google Scholar] [CrossRef]
- Pearce, E.L.; Shen, H. Generation of CD8 T cell memory is regulated by IL-12. J. Immunol. 2007, 179, 2074–2081. [Google Scholar] [CrossRef] [PubMed]
- Agnello, D.; Lankford, C.S.; Bream, J.; Morinobu, A.; Gadina, M.; O’Shea, J.J.; Frucht, D.M. Cytokines and transcription factors that regulate T helper cell differentiation: New players and new insights. J. Clin. Immunol. 2003, 23, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Ben-Sasson, S.Z.; Hu-Li, J.; Quiel, J.; Cauchetaux, S.; Ratner, M.; Shapira, I.; Dinarello, C.A.; Paul, W.E. IL-1 acts directly on CD4 T cells to enhance their antigen-driven expansion and differentiation. Proc. Natl. Acad. Sci. USA 2009, 106, 7119–7124. [Google Scholar] [CrossRef] [PubMed]
- Ben-Sasson, S.Z.; Hogg, A.; Hu-Li, J.; Wingfield, P.; Chen, X.; Crank, M.; Caucheteux, S.; Ratner-Hurevich, M.; Berzofsky, J.A.; Nir-Paz, R.; et al. IL-1 enhances expansion, effector function, tissue localization, and memory response of antigen-specific CD8 T cells. J. Exp. Med. 2013, 210, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Huber, J.P.; Farrar, J.D. Regulation of effector and memory T-cell functions by type I interferon. Immunology 2011, 132, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Martinez, J.; Huang, X.; Yang, Y. Innate immunity against vaccinia virus is mediated by TLR2 and requires TLR-independent production of IFN-β. Blood 2007, 109, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Frenz, T.; Waibler, Z.; Hofmann, J.; Hamdorf, M.; Lantermann, M.; Reizis, B.; Tovey, M.G.; Aichele, P.; Sutter, G.; Kalinke, U. Concomitant type I IFN receptor-triggering of T cells and of DC is required to promote maximal modified vaccinia virus Ankara-induced T-cell expansion. Eur. J. Immunol. 2010, 40, 2769–2777. [Google Scholar] [CrossRef] [PubMed]



© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holgado, M.P.; Falivene, J.; Maeto, C.; Amigo, M.; Pascutti, M.F.; Vecchione, M.B.; Bruttomesso, A.; Calamante, G.; Del Médico-Zajac, M.P.; Gherardi, M.M. Deletion of A44L, A46R and C12L Vaccinia Virus Genes from the MVA Genome Improved the Vector Immunogenicity by Modifying the Innate Immune Response Generating Enhanced and Optimized Specific T-Cell Responses. Viruses 2016, 8, 139. https://doi.org/10.3390/v8050139
Holgado MP, Falivene J, Maeto C, Amigo M, Pascutti MF, Vecchione MB, Bruttomesso A, Calamante G, Del Médico-Zajac MP, Gherardi MM. Deletion of A44L, A46R and C12L Vaccinia Virus Genes from the MVA Genome Improved the Vector Immunogenicity by Modifying the Innate Immune Response Generating Enhanced and Optimized Specific T-Cell Responses. Viruses. 2016; 8(5):139. https://doi.org/10.3390/v8050139
Chicago/Turabian StyleHolgado, María Pía, Juliana Falivene, Cynthia Maeto, Micaela Amigo, María Fernanda Pascutti, María Belén Vecchione, Andrea Bruttomesso, Gabriela Calamante, María Paula Del Médico-Zajac, and María Magdalena Gherardi. 2016. "Deletion of A44L, A46R and C12L Vaccinia Virus Genes from the MVA Genome Improved the Vector Immunogenicity by Modifying the Innate Immune Response Generating Enhanced and Optimized Specific T-Cell Responses" Viruses 8, no. 5: 139. https://doi.org/10.3390/v8050139





