Cloning and Characterization of Sf9 Cell Lamin and the Lamin Conformational Changes during Autographa californica multiple nucleopolyhedrovirus Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Virus
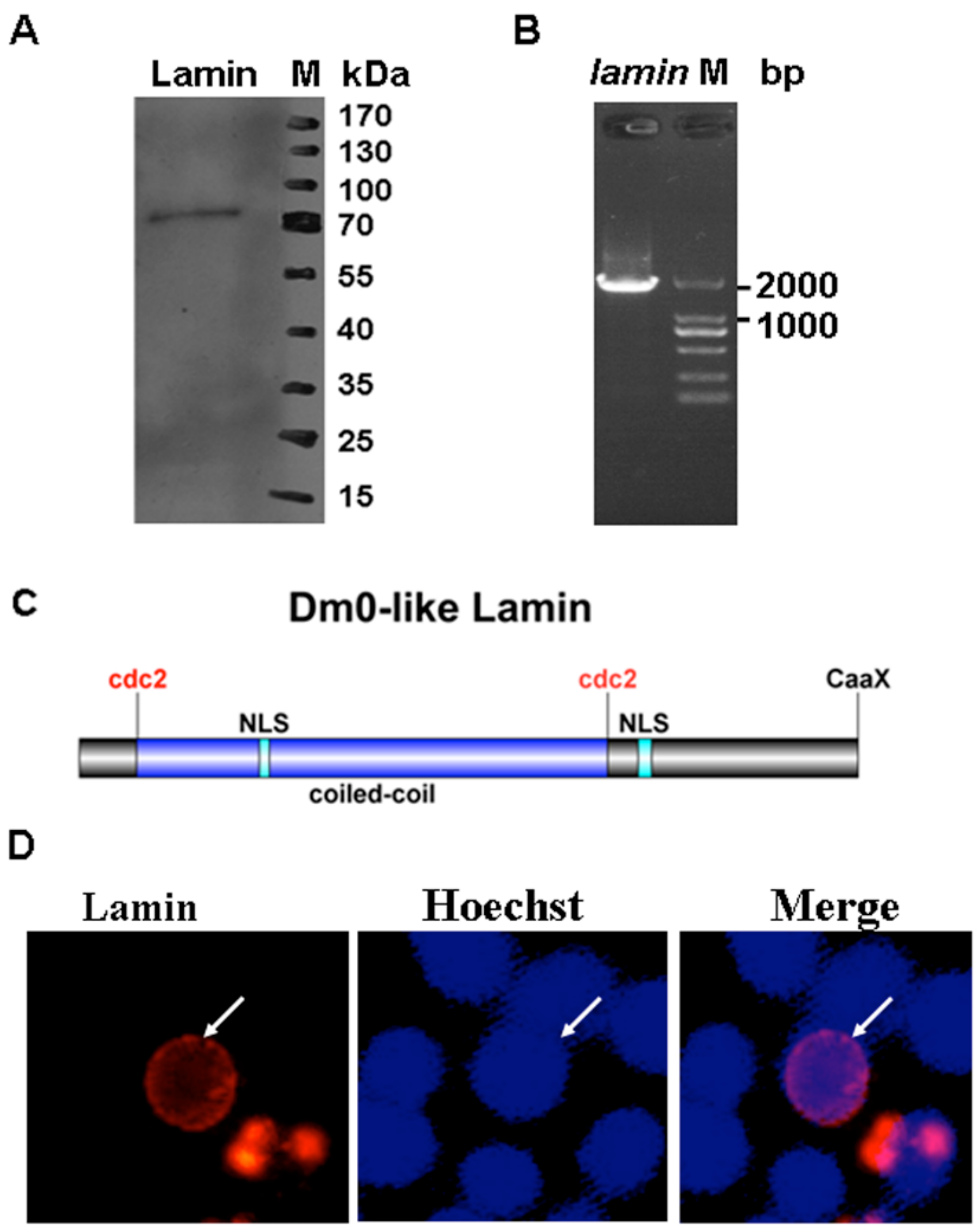
2.2. Reverse Transcriptase Polymerase Chain Reaction (RT-PCR)
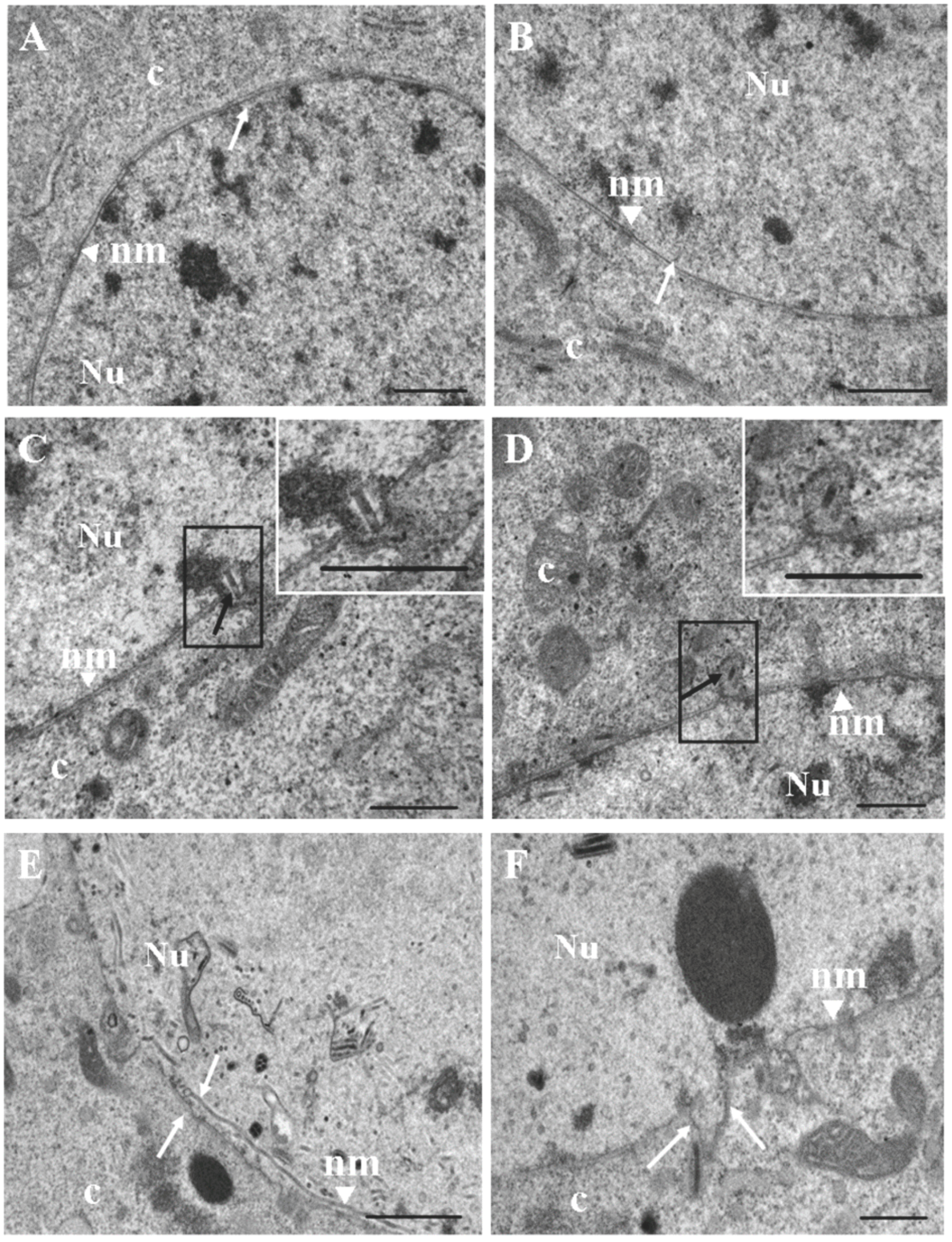
2.3. Transmission Electron Microscopy
2.4. Transfection
2.5. Immunofluorescence
2.6. Western Blotting
3. Results and Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Reddy, K.L.; Zullo, J.M.; Bertolino, E.; Singh, H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 2008, 452, 243–247. [Google Scholar] [CrossRef]
- Taimen, P.; Pfleghaar, K.; Shimi, T.; Möller, D.; Ben-Harush, K.; Erdos, M.R.; Adam, S.A.; Herrmann, H.; Medalia, O.; Collins, F.S.; et al. A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proc. Natl. Acad. Sci. USA 2009, 106, 20788–20793. [Google Scholar] [CrossRef] [PubMed]
- Gruenbaum, Y.; Medalia, O. Lamins: The structure and protein complexes. Curr. Opin. Cell. Biol. 2014, 32, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Adam, S.A.; Goldman, R.D. Insights into the differences between the A- and B-type nuclear lamins. Adv. Enzyme Regul. 2012, 52, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Melcer, S.; Gruenbaum, Y.; Krohne, G. Invertebrate lamins. Exp. Cell Res. 2007, 313, 2157–2166. [Google Scholar] [CrossRef] [PubMed]
- Peter, A.; Stick, R. Evolution of the lamin protein family: What introns can tell. Nucleus 2012, 3, 44–59. [Google Scholar] [CrossRef] [PubMed]
- Dittmer, T.A.; Misteli, T. The lamin protein family. Genome Biol. 2011, 12, 222. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, A.E.; Liang, L.; Baines, J.D. Conformational Changes in the Nuclear Lamina Induced by Herpes Simplex Virus Type 1 Require Genes UL31 and UL34. J. Virol. 2004, 78, 5564–5575. [Google Scholar] [PubMed]
- Park, R.; Baines, J.D. Herpes Simplex Virus Type 1 Infection Induces Activation and Recruitment of Protein Kinase C to the Nuclear Membrane and Increased Phosphorylation of Lamin B. J. Virol. 2006, 80, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Hamirally, S.; Kamil, J.P.; Ndassa-Colday, Y.M.; Lin, A.J.; Jahng, W.J.; Baek, M.C.; Noton, S.; Silva, L.A.; Simpson-Holley, M.; Knipe, D.M.; et al. Viral Mimicry of Cdc2/Cyclin-Dependent Kinase 1 Mediates Disruption of Nuclear Lamina during Human Cytomegalovirus Nuclear Egress. PLoS Pathog. 2009, 5, e1000275. [Google Scholar] [CrossRef] [PubMed]
- Camozzi, D.; Pignatelli, S.; Valvo, C.; Lattanzi, G.; Capanni, C.; Dal Monte, P.; Landini, M.P. Remodelling of the nuclear lamina during human cytomegalovirus infection: Role of the viral proteins pUL50 and pUL53. J. Gen. Virol. 2008, 89, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Herniou, E.A.; Arif, B.M.; Becnel, J.J.; Blissard, G.W.; Bonning, B.; Harrison, R.; Jehle, J.A.; Theilmann, D.A.; Vlak, J.M. Baculoviridae. In Virus Taxonomy Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier-Academic Press: San Diego, CA, USA, 2012; pp. 163–173. [Google Scholar]
- Williams, G.V.; Faulkner, P. The Baculoviruses: Cytological Changes and Viral Morphogenesis during Baculovirus Infection; Miller, L.K., Ed.; Plenum Press: New York, NY, USA, 1997; pp. 61–107. [Google Scholar]
- Knudson, D.L.; Harrap, K.A. Replication of a Nuclear Polyhedrosis Virus in a Continuous Cell Culture of Spodoptera frugiperda: Microscopy Study of the Sequence of Events of the Virus Infection. J. Virol. 1976, 17, 254–268. [Google Scholar]
- Nishimura, T.; Favre, D.; Dürrenberger, M.; Michel, M.R.; Ichihara, I.; Koblet, H. Nuclear export of recombinant baculovirus nucleocapsids through small pore or nuclear-pore-like structure in Sf9 cells. Okajimas Folia Anat. Jpn. 1994, 71, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Dai, X.; Theilmann, D.A. Autographa californica Multiple Nucleopolyhedrovirus EXON0 (ORF141) Is Required for Efficient Egress of Nucleocapsids from the Nucleus. J. Virol. 2007, 81, 9859–9869. [Google Scholar] [CrossRef] [PubMed]
- Ke, J.; Wang, J.; Deng, R.; Wang, X. Autographa californica Multiple Nucleopolyhedrovirus ac66 is required for the efficient egress of nucleocapsids from the nucleus, general synthesis of preoccluded virions and occlusion body formation. Virology 2008, 374, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Huang, Z.; Wei, D.; Hu, Z.; Yang, K.; Pang, Y. Identification of Autographa californica Nucleopolyhedrovirus ac93 as a Core Gene and Its Requirement for Intranuclear Microvesicle Formation and Nuclear Egress of Nucleocapsids. J. Virol. 2011, 85, 11664–11674. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Zhou, Y.; Lei, C.; Sun, X. Autographa californica Multiple Nucleopolyhedrovirus orf114 is not essential for virus replication in vitro, but its knockout reduces per os infectivity in vivo. Virus Genes 2012, 45, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Nègre, V.; Hôtelier, T.; Volkoff, A.-N.; Gimenez, S.; Cousserans, F.; Mita, K.; Sabau, X.; Rocher, J.; López-Ferber, M.; et al. SPODOBASE: An EST database for the lepidopteran crop pest Spodoptera. BMC Bioinform. 2006, 7, 322. [Google Scholar] [CrossRef] [PubMed]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic. Acids. Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: the European Molecular Biology Open Software Suite. Trends. Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Lupas, A.; Van Dyke, M.; Stock, J. Predicting coiled coils from protein sequences. Science 1991, 252, 1162–1164. [Google Scholar] [CrossRef] [PubMed]
- Marin, O.; Meggio, F.; Draetta, G.; Pinna, L.A. The consensus sequences for cdc2 kinase and for casein kinase-2 are mutually incompatible A study with peptides derived from the β-subunit of casein kinase-2. FEBS. Lett. 1992, 301, 111–114. [Google Scholar] [CrossRef]
- Goldberg, T.; Hamp, T.; Rost, B. LocTree2 predicts localization for all domains of life. Bioinformatics 2012, 28, i458–i465. [Google Scholar] [CrossRef] [PubMed]
- Beaudouin, J.; Gerlich, D.; Daigle, N.; Eils, R.; Ellenberg, J. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell 2002, 108, 83–96. [Google Scholar] [CrossRef]
- Panorchan, P.; Schafer, B.W.; Wirtz, D.; Yiider, T. Nuclear envelope breakdown requires overcoming the mechanical integrity of the nuclear lamina. J. Biol. Chem. 2004, 279, 43462–43467. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Yuan, M.; Wu, W.; Liu, C.; Yang, K.; Pang, Y. Autographa californica Multiple Nucleopolyhedrovirus ac76 Is Involved in Intranuclear Microvesicle Formation. J. Virol. 2010, 84, 7437–7447. [Google Scholar] [CrossRef] [PubMed]
- Braunagel, S.C.; Summers, M.D. Molecular Biology of the Baculovirus Occlusion-Derived Virus Envelope. Curr. Drug Targets 2007, 8, 1084–1095. [Google Scholar] [CrossRef] [PubMed]
- Stokke, T.; Steen, H.B. Binding of Hoechst 33258 to chromatin in situ. Cytometry 1986, 7, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Carstens, E.B.; Tjia, S.T.; Doerfler, W. Infection of Spodoptera frugiperda cells with Autographa californica nuclear polyhedrosis virus I. Synthesis of intracellular proteins after virus infection. Virology 1979, 99, 386–398. [Google Scholar] [CrossRef]
- Chen, Y.-R.; Zhong, S.; Fei, Z.; Gao, S.; Zhang, S.; Li, Z.; Wang, P.; Blissard, G.W. Transcriptome Responses of the Host Trichoplusia ni to Infection by the Baculovirus Autographa californica Multiple Nucleopolyhedrovirus. J. Virol. 2014, 88, 13781–13797. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Thiem, S. Responses of insect cells to baculovirus infection: Protein synthesis shutdown and apoptosis. J. Virol. 1997, 71, 7866–7872. [Google Scholar] [PubMed]


| Amino Acid Identity (%) | |||||
|---|---|---|---|---|---|
| Sf9 | Bombyx | Drosophila | Mus | Homo | |
| Bombyx | 92 | ||||
| Drosophila | 50 | 50 | |||
| Mus | 33 | 33 | 31 | ||
| Homo | 31 | 30 | 29 | 78 | |
| Xenopus | 20 | 20 | 18 | 41 | 34 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, W.; Wang, H.; Li, X.; Fang, N.; Yang, S.; Liu, H.; Kang, X.; Sun, X.; Ji, S. Cloning and Characterization of Sf9 Cell Lamin and the Lamin Conformational Changes during Autographa californica multiple nucleopolyhedrovirus Infection. Viruses 2016, 8, 126. https://doi.org/10.3390/v8050126
Wei W, Wang H, Li X, Fang N, Yang S, Liu H, Kang X, Sun X, Ji S. Cloning and Characterization of Sf9 Cell Lamin and the Lamin Conformational Changes during Autographa californica multiple nucleopolyhedrovirus Infection. Viruses. 2016; 8(5):126. https://doi.org/10.3390/v8050126
Chicago/Turabian StyleWei, Wenqiang, Hongju Wang, Xiaoya Li, Na Fang, Shili Yang, Hongyan Liu, Xiaonan Kang, Xiulian Sun, and Shaoping Ji. 2016. "Cloning and Characterization of Sf9 Cell Lamin and the Lamin Conformational Changes during Autographa californica multiple nucleopolyhedrovirus Infection" Viruses 8, no. 5: 126. https://doi.org/10.3390/v8050126






