Infectious Salmon Anaemia Virus (ISAV) RNA Binding Protein Encoded by Segment 8 ORF2 and Its Interaction with ISAV and Intracellular Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells, Virus, Cloning and Transfections
2.1.1. Immunostaining and Microscopy
2.1.2. Immunoprecipitation (IP) and SDS-PAGE
2.1.3. Immunoprecipitation, LC-MS and Data Analysis
2.1.4. Computational Analysis
3. Results
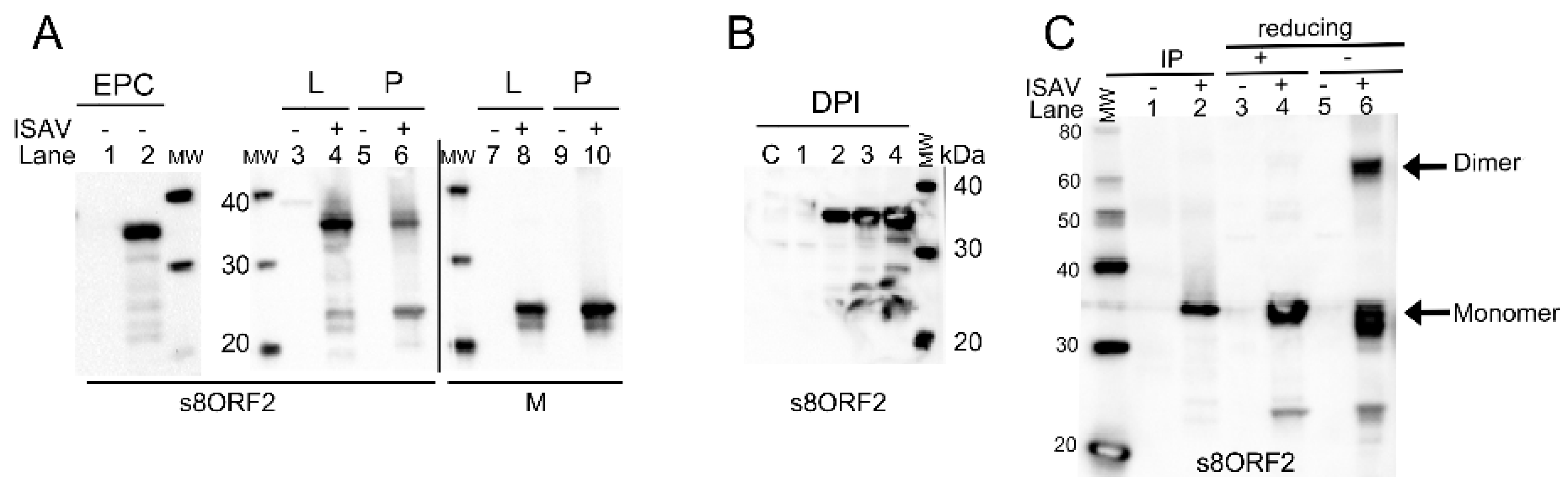
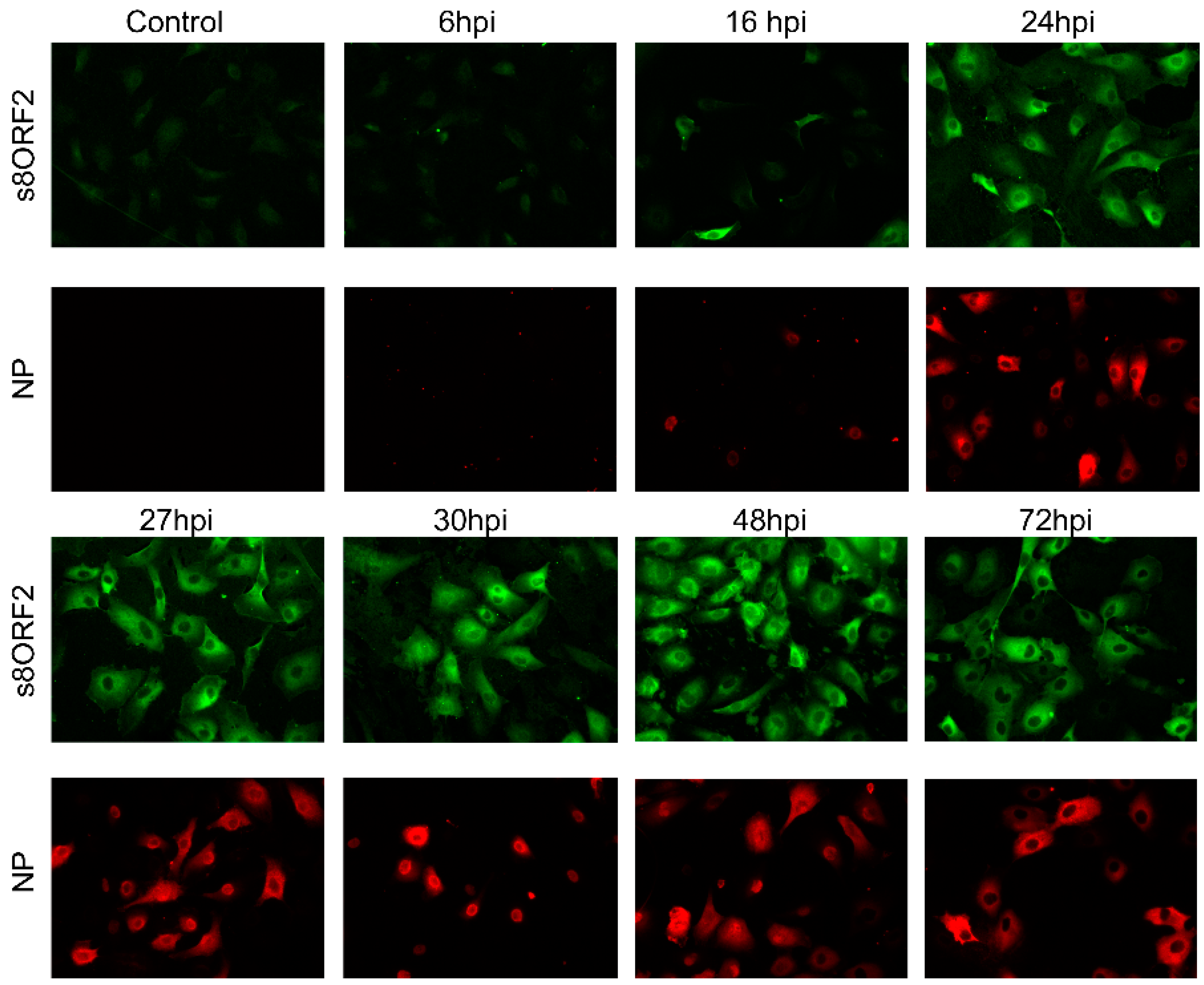
3.1. The s8ORF2 Protein Exists in Different Forms in Infected Cells
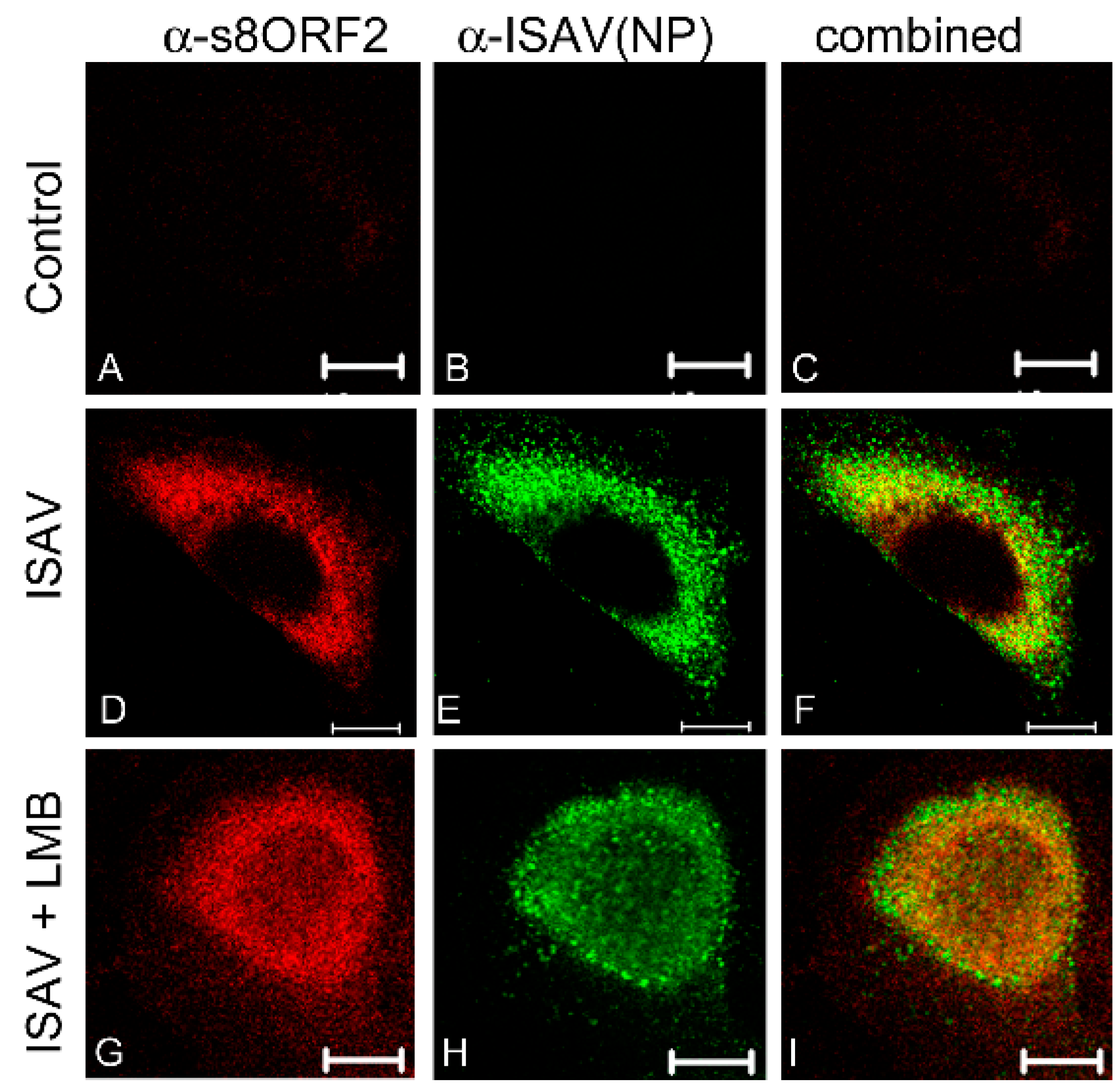
3.1.1. The s8ORF2 Protein Localizes Mainly to the Cytoplasm but is also Present in the Nucleus in ASK Cells
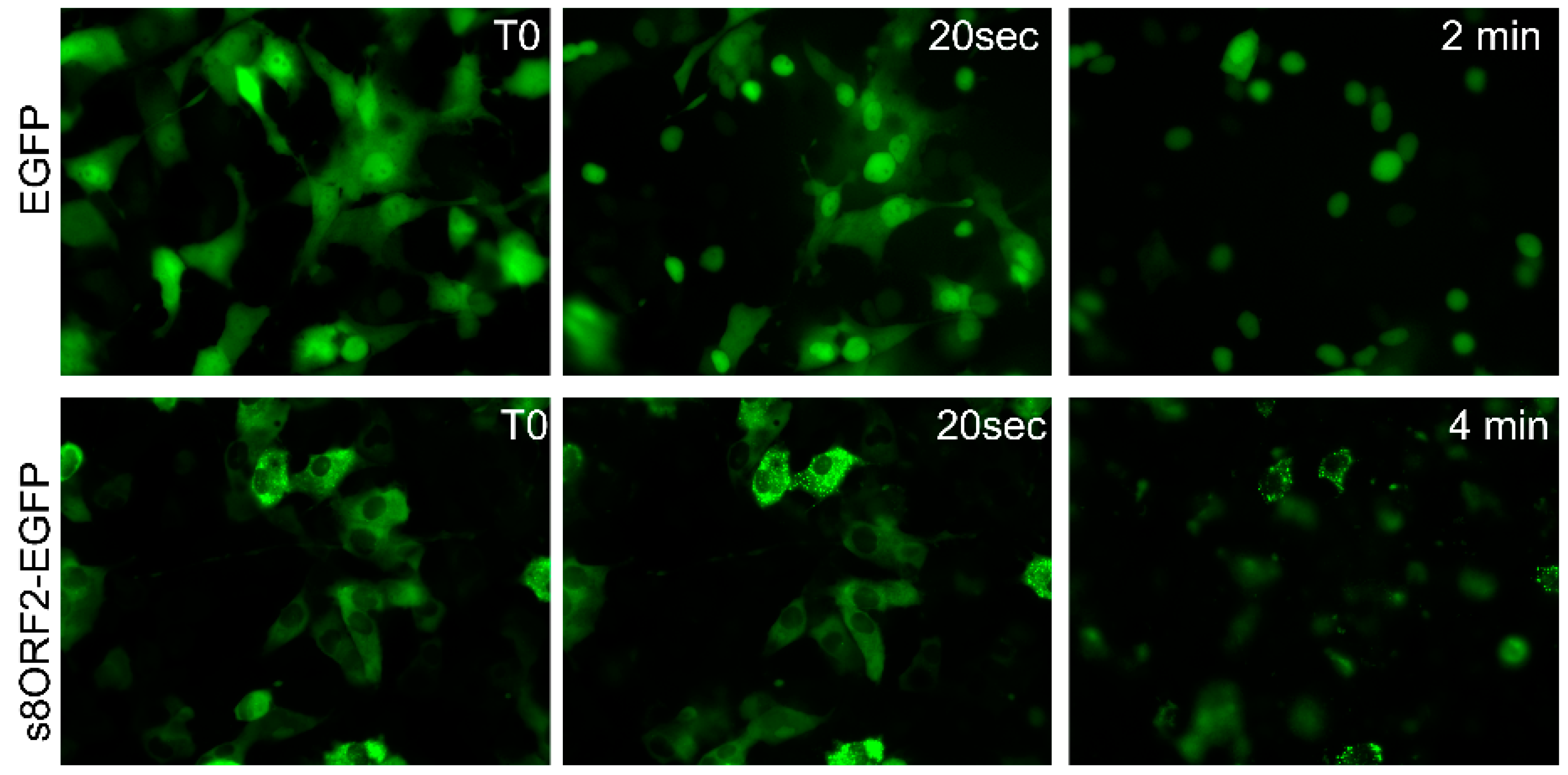
3.1.2. The s8ORF2 Protein is Bound to Intracellular Structures Preventing it from Diffusing Freely out of the Cytoplasm Following Plasma Membrane Permeabilization
3.1.3. The s8ORF2 Protein Interacts with the ISAV Nucleo-, Matrix and Fusion Proteins
3.1.4. In Infected Cells the s8ORF2 Protein Is Conjugated to ISG15 and Ubiquitin
3.1.5. Proteins Associated with Cytoskeleton, Ribosomes, Mitochondria, and Cell Membranes Were Identified in Peptide Analysis Following Immunoprecipitation with Anti-s8ORF2
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Clouthier, S.C.; Rector, T.; Brown, N.E.; Anderson, E.D. Genomic organization of infectious Salmon Anaemia Virus. J. Gen. Virol. 2002, 83, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Falk, K.; Aspehaug, V.; Vlasak, R.; Endresen, C. Identification and Characterization of Viral Structural Proteins of Infectious Salmon Anemia Virus. J. Virol. 2004, 78, 3063–3071. [Google Scholar] [CrossRef] [PubMed]
- Mjaaland, S.; Rimstad, E.; Falk, K.; Dannevig, B.H. Genomic characterization of the virus causing infectious Salmon Anemia in Atlantic Salmon (Salmo Salar L.): An orthomyxo-like virus in a teleost. J. Virol. 1997, 71, 7681–7686. [Google Scholar] [PubMed]
- Biering, E.; Falk, K.; Hoel, E.; Thevarajan, J.; Joerink, M.; Nylund, A.; Endresen, C.; Krossøy, B. Segment 8 encodes a structural protein of infectious Salmon Anaemia Virus (ISAV); the co-linear transcript from segment 7 probably encodes a non-structural or minor structural protein. Dis. Aquat. Organ 2002, 49, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rosado, E.; Markussen, T.; Kileng, O.; Baekkevold, E.S.; Robertsen, B.; Mjaaland, S.; Rimstad, E. Molecular and functional characterization of two infectious salmon anaemia virus (ISAV) proteins with type I interferon antagonizing activity. Virus Res. 2008, 133, 228–238. [Google Scholar] [CrossRef] [PubMed]
- McBeath, A.J.; Collet, B.; Paley, R.; Duraffour, S.; Aspehaug, V.; Biering, E.; Secombes, C.J.; Snow, M. Identification of an interferon antagonist protein encoded by segment 7 of infectious salmon anaemia virus. Virus Res. 2006, 115, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Jensen, I.; Robertsen, B. Effect of double-stranded RNA and interferon on the antiviral activity of atlantic salmon cells against infectious salmon anemia virus and infectious pancreatic necrosis virus. Fish. Shellfish. Immunol. 2002, 13, 221–241. [Google Scholar] [CrossRef] [PubMed]
- Kileng, O.; Brundtland, M.I.; Robertsen, B. Infectious salmon anemia virus is a powerful inducer of key genes of the type I interferon system of Atlantic Salmon, but is not inhibited by interferon. Fish. Shellfish. Immunol. 2007, 23, 378–389. [Google Scholar] [CrossRef] [PubMed]
- McBeath, A.J.; Snow, M.; Secombes, C.J.; Ellis, A.E.; Collet, B. Expression kinetics of interferon and interferon-induced genes in Atlantic Salmon (Salmo Salar) following infection with infectious pancreatic necrosis virus and infectious salmon anaemia virus. Fish. Shellfish. Immunol. 2007, 22, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, H.H.; Schneider, W.M.; Rice, C.M. Interferons and viruses: An evolutionary arms race of molecular interactions. Trends Immunol. 2015, 36, 124–138. [Google Scholar] [CrossRef] [PubMed]
- Morales, D.J.; Lenschow, D.J. The antiviral activities of ISG15. J. Mol. Biol. 2013, 425, 4995–5008. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Q.; Chen, W.; Wang, C. Dynamic regulation of innate immunity by ubiquitin and ubiquitin-like proteins. Cytokine Growth Factor Rev. 2013, 24, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Krug, R.M. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 2001, 20, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Hsiang, T.Y.; Kuo, R.L.; Krug, R.M. ISG15 Conjugation system targets the viral NS1 protein in influenza A virus-infected cells. Proc. Natl. Acad. Sci. USA 2010, 107, 2253–2258. [Google Scholar] [CrossRef] [PubMed]
- Sridharan, H.; Zhao, C.; Krug, R.M. Species specificity of the NS1 protein of influenza B virus: NS1 binds only human and non-human primate ubiquitin-like ISG15 proteins. J. Biol. Chem. 2010, 285, 7852–7856. [Google Scholar] [CrossRef] [PubMed]
- Versteeg, G.A.; Hale, B.G.; van, B.S.; Wolff, T.; Lenschow, D.J.; Garcia-Sastre, A. Species-specific antagonism of host ISGylation by the influenza B virus NS1 protein. J. Virol. 2010, 84, 5423–5430. [Google Scholar] [CrossRef] [PubMed]
- Bodewes, R.; Morick, D.; de, M.G.; Osinga, N.; Bestebroer, T.; van der Vliet, S.; Smits, S.L.; Kuiken, T.; Rimmelzwaan, G.F.; Fouchier, R.A.; et al. Recurring influenza B virus infections in Seals. Emerg. Infect. Dis. 2013, 19, 511–512. [Google Scholar] [CrossRef] [PubMed]
- Osterhaus, A.D.; Rimmelzwaan, G.F.; Martina, B.E.; Bestebroer, T.M.; Fouchier, R.A. Influenza B virus in seals. Science 2000, 288, 1051–1053. [Google Scholar] [CrossRef] [PubMed]
- Ramis, A.J.; van, R.D.; van de Bildt, M.W.; Osterhaus, A.; Kuiken, T. Influenza A and B virus attachment to respiratory tract in marine mammals. Emerg. Infect. Dis. 2012, 18, 817–820. [Google Scholar] [CrossRef] [PubMed]
- Svingerud, T.; Holand, J.K.; Robertsen, B. Infectious salmon anemia virus (ISAV) replication is transiently inhibited by Atlantic Salmon type I interferon in cell culture. Virus Res. 2013, 177, 163–170. [Google Scholar] [CrossRef] [PubMed]
- McBeath, A.J.; Ho, Y.M.; Aamelfot, M.; Hall, M.; Christiansen, D.H.; Markussen, T.; Falk, K.; Matejusova, I. Low virulent infectious Salmon Anaemia Virus (ISAV) replicates and initiates the immune response earlier than a highly virulent virus in Atlantic Salmon Gills. Vet. Res. 2014, 45. [Google Scholar] [CrossRef] [PubMed]
- Devold, M.; Krossøy, B.; Aspehaug, V.; Nylund, A. Use of RT-PCR for diagnosis of infectious Salmon Anaemia Virus (ISAV) in Carrier Sea trout salmo trutta after experimental infection. Dis. Aquat. Organ 2000, 40, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Dannevig, B.H.; Falk, K.; Namork, E. Isolation of the causal virus of infectious Salmon Anaemia (ISA) in a long-term cell line from Atlantic Salmon head kidney. J. Gen. Virol. 1995, 76, 1353–1359. [Google Scholar] [CrossRef] [PubMed]
- Aspehaug, V.; Mikalsen, A.B.; Snow, M.; Biering, E.; Villoing, S. Characterization of the infectious Salmon Anemia Virus fusion protein. J. Virol. 2005, 79, 12544–12553. [Google Scholar] [CrossRef] [PubMed]
- Røkenes, T.P.; Larsen, R.; Robertsen, B. Atlantic Salmon ISG15: Expression and conjugation to cellular proteins in response to interferon, double-stranded RNA and virus infections. Mol. Immunol. 2007, 44, 950–959. [Google Scholar] [CrossRef] [PubMed]
- Ramly, R.B.; Olsen, C.M.; Braaen, S.; Rimstad, E. Infectious Salmon Anaemia Virus nuclear export protein is encoded by a spliced gene product of genomic segment 7. Virus Res. 2013, 177, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Koehler, C.J.; Strozynski, M.; Kozielski, F.; Treumann, A.; Thiede, B. Isobaric peptide termini labeling for MS/MS-based quantitative proteomics. J. Proteome. Res. 2009, 8, 4333–4341. [Google Scholar] [CrossRef] [PubMed]
- ExPASy Bioinformatics Resources Portal Compute pI/Mw tool. Available online: http://web.expasy.org/compute_pi/ (accessed on 27 April 2015).
- SignalP 4.1 Server. Available online: http://www.cbs.dtu.dk/services/SignalP/ (accessed on 19 May 2015).
- ExPASy Bioinformatics Resources Portal TMpred. Available online: http://www.ch.embnet.org/software/TMPRED_form.html (accessed on 19 May 2015).
- NetNGlyc 1.0 Server. Available online: http://www.cbs.dtu.dk/services/NetNGlyc/ (accessed on 19 May 2015).
- The PSIPRED Protein Sequence Analysis Workbench. Available online: http://bioinf.cs.ucl.ac.uk/psipred/ (accessed on 5 June 2015).
- ExPASy Bioinformatics Resources Portal ProtScale. Available online: http://web.expasy.org/protscale/ (accessed on 5 June 2015).
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Gravy Calculator. Available online: http://www.gravy-calculator.de/ (accessed on 19 May 2015).
- EMBOSS explorer epestfind. Available online: http://emboss.bioinformatics.nl/cgi-bin/emboss/epestfind (accessed on 30 April 2015).
- NoD Nucleolar Localization Sequence Detector. Available online: http://www.compbio.dundee.ac.uk/www-nod/ (accessed on 5 June 2015).
- Rogers, S.; Wells, R.; Rechsteiner, M. Amino acid sequences common to rapidly degraded proteins: The PEST hypothesis. Science 1986, 234, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Alai, M.M.; Gallo, M.; Salame, M.; Wetzler, D.E.; McBride, A.A.; Paci, M.; Cicero, D.O.; de Prat-Gay, G. Molecular basis for phosphorylation-dependent, PEST-mediated protein turnover. Structure 2006, 14, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Hericourt, F.; Blanc, S.; Redeker, V.; Jupin, I. Evidence for phosphorylation and ubiquitinylation of the turnip yellow mosaic virus RNA-dependent RNA polymerase domain expressed in a baculovirus-insect cell system. Biochem. J. 2000, 349, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Rechsteiner, M.; Rogers, S.W. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 1996, 21, 267–271. [Google Scholar] [CrossRef]
- Melen, K.; Kinnunen, L.; Fagerlund, R.; Ikonen, N.; Twu, K.Y.; Krug, R.M.; Julkunen, I. Nuclear and nucleolar targeting of influenza A virus NS1 protein: Striking differences between different virus subtypes. J. Virol. 2007, 81, 5995–6006. [Google Scholar] [CrossRef] [PubMed]
- Ayllon, J.; Garcia-Sastre, A. The NS1 protein: A multitasking virulence factor. Curr. Top. Microbiol. Immunol. 2015, 386, 73–107. [Google Scholar] [PubMed]
- Nemeroff, M.E.; Qian, X.Y.; Krug, R.M. The influenza-virus Ns1 protein forms multimers in-vitro and in-vivo. Virology 1995, 212, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Burkhard, P.; Stetefeld, J.; Strelkov, S.V. Coiled coils: A highly versatile protein folding motif. Trends Cell Biol. 2001, 11, 82–88. [Google Scholar] [CrossRef]
- Giannakopoulos, N.V.; Luo, J.K.; Papov, V.; Zou, W.; Lenschow, D.J.; Jacobs, B.S.; Borden, E.C.; Li, J.; Virgin, H.W.; Zhang, D.E. Proteomic identification of proteins conjugated to ISG15 in mouse and human cells. Biochem. Biophys. Res. Commun. 2005, 336, 496–506. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Collins, M.N.; Hsiang, T.Y.; Krug, R.M. Interferon-induced ISG15 pathway: An ongoing virus-host battle. Trends Microbiol. 2013, 21, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Geiss-Friedlander, R.; Melchior, F. Concepts in sumoylation: A decade on. Nat. Rev. Mol. Cell Biol. 2007, 8, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Kumakura, M.; Kawaguchi, A.; Nagata, K. Actin-myosin network is required for proper assembly of influenza virus particles. Virology 2015, 476, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Meister, M.; Tikkanen, R. Endocytic trafficking of membrane-bound cargo: A flotillin point of view. Membranes 2014, 4, 356–371. [Google Scholar] [CrossRef] [PubMed]
- Berri, F.; Haffar, G.; Le, V.B.; Sadewasser, A.; Paki, K.; Lina, B.; Wolff, T.; Riteau, B. Annexin V incorporated into influenza virus particles inhibits Gamma interferon signaling and promotes viral replication. J. Virol. 2014, 88, 11215–11228. [Google Scholar] [CrossRef] [PubMed]
- Shaw, M.L.; Stone, K.L.; Colangelo, C.M.; Gulcicek, E.E.; Palese, P. Cellular proteins in influenza virus particles. PLoS. Pathog. 2008, 4, e1000085. [Google Scholar] [CrossRef] [PubMed]
- Geller, R.; Taguwa, S.; Frydman, J. Broad action of Hsp90 as a host chaperone required for viral replication. Biochim. Biophys. Acta 2012, 1823, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P. Recruitment of Hsp70 chaperones: A crucial part of viral survival strategies. Rev. Physiol. Biochem. Pharmacol. 2005, 153, 1–46. [Google Scholar] [PubMed]
- Radhakrishnan, A.; Yeo, D.; Brown, G.; Myaing, M.Z.; Iyer, L.R.; Fleck, R.; Tan, B.H.; Aitken, J.; Sanmun, D.; Tang, K.; et al. Protein analysis of purified respiratory syncytial virus particles reveals an important role for heat shock protein 90 in virus particle assembly. Mol. Cell Proteomics 2010, 9, 1829–1848. [Google Scholar] [CrossRef] [PubMed]
- Jagdeo, J.M.; Dufour, A.; Fung, G.; Luo, H.; Kleifeld, O.; Overall, C.M.; Jan, E. Heterogeneous nuclear ribonucleoprotein M facilitates enterovirus infection. J. Virol. 2015, 89, 7064–7078. [Google Scholar] [CrossRef] [PubMed]
- Naji, S.; Ambrus, G.; Cimermancic, P.; Reyes, J.R.; Johnson, J.R.; Filbrandt, R.; Huber, M.D.; Vesely, P.; Krogan, N.J.; Yates, J.R., III; Saphire, A.C.; Gerace, L. Host cell interactome of HIV-1 rev includes RNA helicases involved in multiple facets of virus production. Mol. Cell Proteomics 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Ge, X.; Wang, X.; Liu, A.; Guo, X.; Zhou, L.; Yu, K.; Yang, H. The DEAD-box RNA helicase 5 positively regulates the replication of porcine reproductive and respiratory syndrome virus by interacting with viral Nsp9 in Vitro. Virus Res. 2015, 195, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ge, L.L.; Li, P.P.; Wang, Y.; Sun, M.X.; Huang, L.; Ishag, H.; Di, D.D.; Shen, Z.Q.; Fan, W.X.; Mao, X. The DEAD-box RNA helicase DDX5 acts as a positive regulator of Japanese Encephalitis Virus replication by binding to viral 3′UTR. Antivir. Res. 2013, 100, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Zamarin, D.; Garcia-Sastre, A.; Xiao, X.; Wang, R.; Palese, P. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS. Pathog. 2005, 1, e4. [Google Scholar] [CrossRef] [PubMed]
- Schiøtz, B.L.; Roos, N.; Rishovd, A.L.; Gjøen, T. Formation of autophagosomes and redistribution of LC3 upon in vitro infection with infectious Salmon Anemia Virus. Virus Res. 2010, 151, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Schiøtz, B.L.; Baekkevold, E.S.; Poulsen, L.C.; Mjaaland, S.; Gjøen, T. Analysis of host- and strain dependent cell death responses during infectious Salmon Anemia Virus infection in Vitro. Virol. J. 2009, 6, 91. [Google Scholar] [CrossRef] [PubMed]
- Schiøtz, B.L.; Jørgensen, S.M.; Rexroad, C.; Gjøen, T.; Krasnov, A. Transcriptomic analysis of responses to infectious Salmon Anemia Virus infection in macrophage-like cells. Virus Res. 2008, 136, 65–74. [Google Scholar] [CrossRef] [PubMed]



| Protein Band | a Data-Base | Protein Name | NCBI Accession No. | Mw (kDa) | Spectrum Count | Function |
|---|---|---|---|---|---|---|
| 1 | S. salar | Myosin-9 | 224613261 | 60 | 13 | Actin binding |
| S. salar | Eukaryotic translation initiation factor 3 subunit | 223647896 | 115 | 3 | Translation | |
| 2 | S. salar | Myosin-9 | 224613261 | 60 | 6 | Actin binding |
| O. mykiss | Actin beta | 185132289 | 42 | 9 | Actin | |
| O. mykiss | Heat shock 90 KDa protein 1 beta isoform | 185132161 | 83 | 2 | Chaperone | |
| 3 | S. salar | Myosin-9 | 224613261 | 60 | 7 | Actin binding |
| O. mykiss | Heat shock 90 KDa protein 1 beta isoform | 185132161 | 83 | 2 | Chaperone | |
| 4 | O. mykiss | Heat shock 90 KDa protein 1 beta isoform | 185132161 | 83 | 16 | Chaperone |
| S. salar | Mitochondrial inner membrane protein | 209153972 | 79 | 13 | Transport | |
| S. salar | Myosin-9 | 224613261 | 60 | 11 | Actin binding | |
| S. salar | Annexin A1 | 213510942 | 38 | 3 | Multifunctional | |
| Virus | NP protein | 313744891 | 68 | 2 | ISAV | |
| 5 | Virus | NP protein | 313744891 | 68 | 14 | ISAV |
| S. salar | Myosin-9 | 224613261 | 60 | 12 | Actin binding | |
| S. salar | Heat shock cognate 71 kDa protein | 213514058 | 71 | 3 | Chaperone | |
| S. salar | Myelin expression factor 2 | 291190830 | 65 | 3 | Transcription | |
| 6 | S. salar | Flotillin 1 | 213511228 | 47 | 7 | Scaffolding protein |
| S. salar | Myosin-9 | 224613261 | 60 | 5 | Actin binding | |
| S. salar | Flotillin-2a | 213514074 | 47 | 2 | Scaffolding protein | |
| Virus | S8ORF2 protein | 313754903 | 27 | 3 | ISAV | |
| Virus | NP protein | 313744891 | 68 | 2 | ISAV | |
| 7 | Virus | NP protein | 313744891 | 68 | 70 | ISAV |
| S. salar | Myelin expression factor 2 | 291190830 | 65 | 8 | Transcription | |
| S. salar | Heat shock cognate 71 kDa protein | 213514058 | 71 | 7 | Chaperone | |
| S. salar | Myosin-9 | 224613261 | 60 | 6 | Actin binding | |
| S. salar | Heterogeneous nuclear ribonucleoprotein M | 213512325 | 72 | 4 | Pre-mRNA binding | |
| S. salar | Probable ATP-dependent RNA helicase DDX5 | 223649022 | 68 | 4 | mRNA processing | |
| S. salar | Annexin A6 | 213514676 | 75 | 2 | Multifunctional | |
| S. salar | Syncoilin | 213512082 | 64 | 2 | Intermediate filament | |
| 8 | S. salar | Myosin-9 | 224613261 | 60 | 6 | Actin binding |
| S. salar | Galectin-9 | 209733430 | 38 | 4 | Carbohydrate binding | |
| Virus | NP protein | 313744891 | 68 | 3 | ISAV | |
| S. salar | Annexin A1 | 213510942 | 38 | 3 | Multifunctional | |
| S. salar | Capping protein (Actin filament) muscle Z-line α 2 | 213510872 | 33 | 3 | Actin binding | |
| S. salar | F-actin-capping protein subunit-1 | 223647378 | 33 | 3 | Actin binding | |
| Virus | S8ORF2 protein | 313754903 | 27 | 2 | ISAV | |
| S. salar | Voltage-dependent anion channel 3 | 213511881 | 30 | 2 | Transport | |
| 9 | Virus | S8ORF2 protein | 313754903 | 27 | 13 | ISAV |
| S. salar | Capping protein (actin filament) muscle Z-line β | 197631853 | 31 | 3 | Actin binding | |
| Virus | NP protein | 313744891 | 68 | 2 | ISAV | |
| O. mykiss | Actin β | 185132289 | 42 | 2 | Actin | |
| 10 | S. salar | Tropomyosin α-3 chain | 218505649 | 29 | 16 | Actin binding |
| O. mykiss | Actin β | 185132289 | 42 | 15 | Actin | |
| S. salar | Tropomyosin α-3 chain | 223647762 | 28 | 15 | Actin binding | |
| S. salar | Tropomyosin α-4 chain | 213515262 | 29 | 10 | Actin binding | |
| S. salar | Voltage-dependent anion channel 2-2 | 197632613 | 30 | 8 | Transport | |
| S. salar | 40S ribosomal protein S3a | 213514340 | 30 | 7 | Ribosomal protein | |
| S. salar | Ribosomal protein S3-1 | 197632569 | 27 | 6 | Ribosomal protein | |
| Virus | s8ORF2 protein | 313754903 | 27 | 5 | ISAV | |
| S. salar | Myosin-9 | 224613264 | 60 | 5 | Actin binding | |
| S. salar | Ribosomal protein L7a | 198285627 | 30 | 5 | Ribosomal protein | |
| Virus | Tyrosine-protein kinase transforming protein Fgr | 125357 | 62 | 4 | Kinase, transferase | |
| S. salar | Voltage-dependent anion-selective channel protein 2 | 209154650 | 34 | 4 | Transport | |
| S. salar | THO complex subunit 4 | 209733738 | 28 | 3 | mRNA processing | |
| S. salar | 60S ribosomal protein L6 | 213511212 | 30 | 2 | Ribosomal protein | |
| S. salar | Annexin A5 | 213514536 | 35 | 2 | Multifunctional | |
| S. salar | Capping protein (actin filament) muscle Z-line β | 197631853 | 31 | 2 | Actin binding | |
| S. salar | Voltage-dependent anion-selective channel protein 1 | 209156112 | 31 | 2 | Transport |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olsen, C.M.; Markussen, T.; Thiede, B.; Rimstad, E. Infectious Salmon Anaemia Virus (ISAV) RNA Binding Protein Encoded by Segment 8 ORF2 and Its Interaction with ISAV and Intracellular Proteins. Viruses 2016, 8, 52. https://doi.org/10.3390/v8020052
Olsen CM, Markussen T, Thiede B, Rimstad E. Infectious Salmon Anaemia Virus (ISAV) RNA Binding Protein Encoded by Segment 8 ORF2 and Its Interaction with ISAV and Intracellular Proteins. Viruses. 2016; 8(2):52. https://doi.org/10.3390/v8020052
Chicago/Turabian StyleOlsen, Christel M., Turhan Markussen, Bernd Thiede, and Espen Rimstad. 2016. "Infectious Salmon Anaemia Virus (ISAV) RNA Binding Protein Encoded by Segment 8 ORF2 and Its Interaction with ISAV and Intracellular Proteins" Viruses 8, no. 2: 52. https://doi.org/10.3390/v8020052






