Structures and Functions of Pestivirus Glycoproteins: Not Simply Surface Matters
Abstract
:1. Introduction
2. Structures of the Pestivirus Glycoproteins
2.1. Crystal Structures of the Glycoproteins
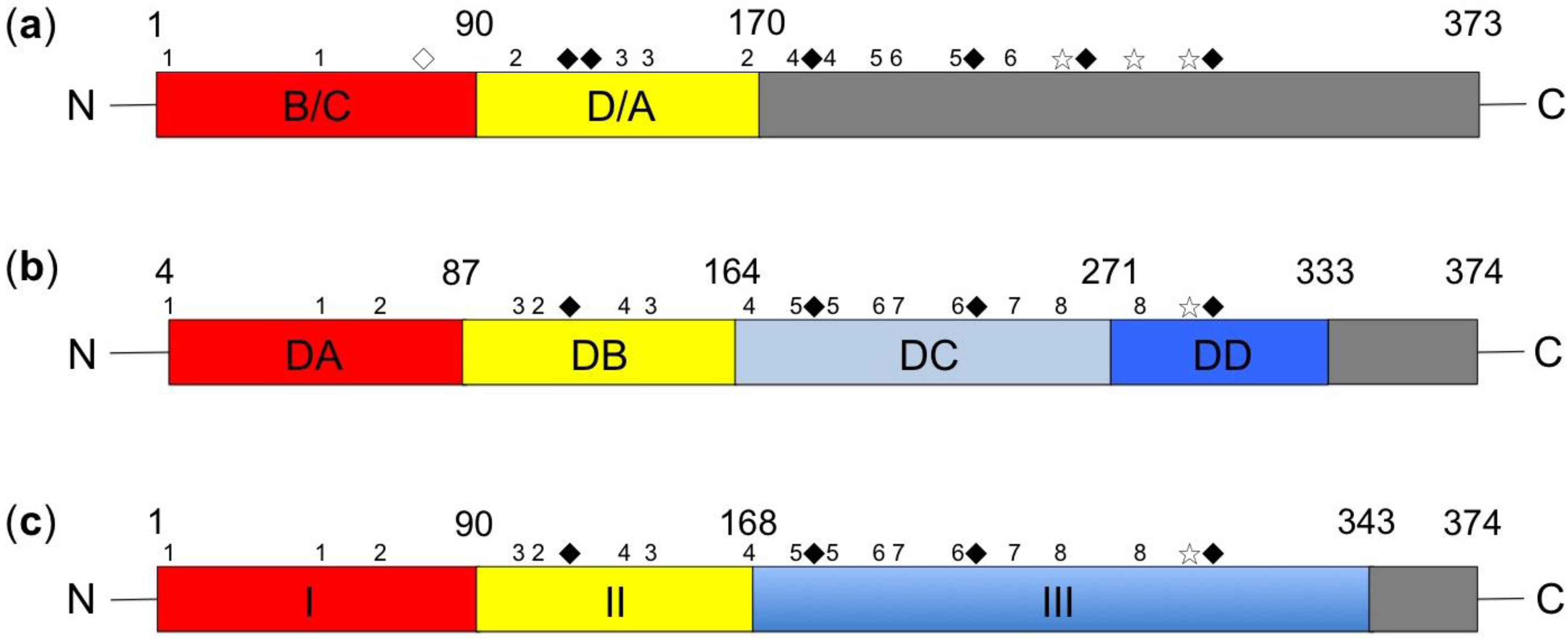
2.2. Intramolecular Disulfide Linkage and Dimerization of Glycoproteins
2.3. N- and O-Linked Glycosylation of Glycoproteins
2.4. Antigenic Structure and Epitopes of Glycoproteins
3. Functions of Glycoproteins during the Pestivirus Life Cycle
| Category | Functions | References |
|---|---|---|
| Interactions with cells | Attachment: Erns and E2 | [72] |
| Entry: E1 and E2 | [36,72,73] | |
| Cultured cell tropism and host specificity: E2 | [74,75,76] | |
| Interactions with cellular receptors: Erns (heparan sulfate, laminin receptor) and E2 (CD46, heparan sulfate) | [77,78,79,80,81,82,83,84] | |
| Interactions with cellular proteins: E2 | [85,86,87,88] | |
| Fusion: E2 (CSFV) and E1 (BVDV) | [22,29,30,89,90,91,92,93,94] | |
| Endocytosis: Erns | [91,92,95] | |
| Autophagy: Erns and E2 | [96,97,98] | |
| Interactions with other viral proteins | Dimerization: E1-E2 heterodimer (major), Erns homodimer, and E2 homodimer | [5] |
| Virion packaging and assembly: E2 homodimer early and then E1-E2 heterodimer later | [99] | |
| Functions in pathogenesis | Interactions with receptors to determine cell tropism and pathogenicity | [74,75] |
| Eliciting host humoral immunity: E2 induces the major neutralization antibody, and Erns induces the second neutralization antibody | [8,9,10] | |
| Eliciting host cellular immunity: E2 is the target of CTL, and Erns and E1 also have roles | [100,101,102] | |
| Evasion from immunity: RNase activity of Erns induces apoptosis and inhibits IFN synthesis; Erns and E2 are responsible for positive selection | [43,103,104,105,106,107,108,109,110,111] | |
| Virulence: Erns, E1 and E2 | [21,33,35,39,40,41,112,113,114,115,116,117,118,119,120,121] |
3.1. Interactions with Cells
3.2. Interactions with Other Proteins within Virions
3.3. Interactions with the Host
4. The Roles of Viral Glycoproteins in Pathogenicity in Animals or Cells
4.1. Genetic Basis of Pestivirus Virulence
4.2. Antigenic Differences Influence the Efficacy of E2 Subunit Vaccines
4.3. Selection Pressure-Driven Mutations Determine the Appropriateness of Vaccines
5. Concluding Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Simmonds, P.; Becher, P.; Collett, M.; Gould, E.; Heinz, F.; Meyers, G.; Monath, T.; Pletnev, A.; Rice, C.; Stiasny, K.; et al. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Eds.; Elsevier Academic Press: San Diego, CA, USA, 2011; pp. 1003–1020. [Google Scholar]
- Lindenbach, B.D.; Thiel, H.J.; Rice, C.M. Flaviviridae: The viruses and their replication. In Fields Virology, 5th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; pp. 1101–1152. [Google Scholar]
- Becher, P.; Avalos Ramirez, R.; Orlich, M.; Cedillo Rosales, S.; Konig, M.; Schweizer, M.; Stalder, H.; Schirrmeier, H.; Thiel, H.J. Genetic and antigenic characterization of novel pestivirus genotypes: Implications for classification. Virology 2003, 311, 96–104. [Google Scholar] [CrossRef]
- Meyers, G.; Thiel, H.J. Molecular characterization of pestiviruses. Adv. Virus Res. 1996, 47, 53–118. [Google Scholar] [PubMed]
- Thiel, H.J.; Stark, R.; Weiland, E.; Rümenapf, T.; Meyers, G. Hog cholera virus: Molecular composition of virions from a pestivirus. J. Virol. 1991, 65, 4705–4712. [Google Scholar] [PubMed]
- Rümenapf, T.; Unger, G.; Strauss, J.H.; Thiel, H.J. Processing of the envelope glycoproteins of pestiviruses. J. Virol. 1993, 67, 3288–3294. [Google Scholar] [PubMed]
- Bintintan, I.; Meyers, G. A new type of signal peptidase cleavage site identified in an RNA virus polyprotein. J. Biol. Chem. 2010, 285, 8572–8584. [Google Scholar] [CrossRef] [PubMed]
- Weiland, E.; Stark, R.; Haas, B.; Rümenapf, T.; Meyers, G.; Thiel, H.J. Pestivirus glycoprotein which induces neutralizing antibodies forms part of a disulfide-linked heterodimer. J. Virol. 1990, 64, 3563–3569. [Google Scholar] [PubMed]
- König, M.; Lengsfeld, T.; Pauly, T.; Stark, R.; Thiel, H.J. Classical swine fever virus: Independent induction of protective immunity by two structural glycoproteins. J. Virol. 1995, 69, 6479–6486. [Google Scholar] [PubMed]
- Weiland, E.; Ahl, R.; Stark, R.; Weiland, F.; Thiel, H.J. A second envelope glycoprotein mediates neutralization of a pestivirus, hog cholera virus. J. Virol. 1992, 66, 3677–3682. [Google Scholar] [PubMed]
- Hulst, M.M.; Himes, G.; Newbigin, E.; Moormann, R.J. Glycoprotein E2 of classical swine fever virus: Expression in insect cells and identification as a ribonuclease. Virology 1994, 200, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Schneider, R.; Unger, G.; Stark, R.; Schneider-Scherzer, E.; Thiel, H.J. Identification of a structural glycoprotein of an RNA virus as a ribonuclease. Science 1993, 261, 1169–1171. [Google Scholar] [CrossRef] [PubMed]
- Fetzer, C.; Tews, B.A.; Meyers, G. The carboxy-terminal sequence of the pestivirus glycoprotein Erns represents an unusual type of membrane anchor. J. Virol. 2005, 79, 11901–11913. [Google Scholar] [CrossRef] [PubMed]
- Tews, B.A.; Meyers, G. The pestivirus glycoprotein Erns is anchored in plane in the membrane via an amphipathic helix. J. Biol. Chem. 2007, 282, 32730–32741. [Google Scholar] [CrossRef] [PubMed]
- Aberle, D.; Muhle-Goll, C.; Bürck, J.; Wolf, M.; Reißer, S.; Luy, B.; Wenzel, W.; Ulrich, A.S.; Meyers, G. Structure of the membrane anchor of pestivirus glycoprotein Erns, a long tilted amphipathic helix. PLoS Pathog. 2014, 10, e1003973. [Google Scholar] [CrossRef] [PubMed]
- Burrack, S.; Aberle, D.; Bürck, J.; Ulrich, A.S.; Meyers, G. A new type of intracellular retention signal identified in a pestivirus structural glycoprotein. FASEB J. 2012, 26, 3292–3305. [Google Scholar] [CrossRef] [PubMed]
- Hulst, M.M.; van Gennip, H.G.; Moormann, R.J. Passage of classical swine fever virus in cultured swine kidney cells selects virus variants that bind to heparan sulfate due to a single amino acid change in envelope protein Erns. J. Virol. 2000, 74, 9553–9561. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.; McCauley, J.W. Identification of the glycosaminoglycan binding site on the glycoprotein Erns of bovine viral diarrhoea virus by site directed mutagenesis. J. Gen. Virol. 2002, 83, 2153–2159. [Google Scholar] [PubMed]
- Krey, T.; Bontems, F.; Vonrhein, C.; Vaney, M.C.; Bricogne, G.; Rümenapf, T.; Rey, F.A. Crystal structure of the pestivirus envelope glycoprotein Erns and mechanistic analysis of its ribonuclease activity. Structure 2012, 20, 862–873. [Google Scholar] [CrossRef] [PubMed]
- Hulst, M.; Panoto, F.; Hoekman, A.; van Gennip, H.; Moormann, R. Inactivation of the RNase activity of glycoprotein Erns of classical swine fever virus results in a cytopathogenic virus. J. Virol. 1998, 72, 151–157. [Google Scholar] [PubMed]
- Meyers, G.; Saalmüller, A.; Büttner, M. Mutations abrogating the RNase activity in glycoprotein Erns of the pestivirus classical swine fever virus lead to virus attenuation. J. Virol. 1999, 73, 10224–10235. [Google Scholar] [PubMed]
- Wang, J.; Li, Y.; Modis, Y. Structural models of the membrane anchors of envelope glycoproteins E1 and E2 from pestiviruses. Virology 2014, 454–455, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Wensvoort, G. Topographical and functional mapping of epitopes on hog cholera virus with monoclonal antibodies. J. Gen. Virol. 1989, 70, 2865–2876. [Google Scholar] [CrossRef] [PubMed]
- Wensvoort, G.; Boonstra, J.; Bodzinga, B.G. Immunoaffinity purification and characterization of the envelope protein E1 of hog cholera virus. J. Gen. Virol. 1990, 71, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.Y.; Huang, C.C.; Lin, Y.J.; Deng, M.C.; Chen, H.C.; Tsai, C.H.; Chang, W.M.; Wang, F.I. Antigenic domains analysis of classical swine fever virus E2 glycoprotein by mutagenesis and conformation-dependent monoclonal antibodies. Virus Res. 2010, 149, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.Y.; Huang, C.C.; Deng, M.C.; Huang, Y.L.; Lin, Y.J.; Liu, H.M.; Lin, Y.L.; Wang, F.I. Identification of conformational epitopes and antigen-specific residues at the D/A domains and the extramembrane C-terminal region of E2 glycoprotein of classical swine fever virus. Virus Res. 2012, 168, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Van Rijn, P.A.; Miedema, G.K.W.; Wensvoort, G.; van Gennip, H.G.P.; Moormann, R.J.M. Antigenic structure of envelope glycoprotein E1 of hog cholera virus. J. Virol. 1994, 68, 3934–3942. [Google Scholar] [PubMed]
- Huang, Y.L.; Deng, M.C.; Wang, F.I.; Huang, C.C.; Chang, C.Y. The challenges of classical swine fever control: Modified live and E2 subunit vaccines. Virus Res. 2014, 179, 1–11. [Google Scholar] [CrossRef] [PubMed]
- El Omari, K.; Iourin, O.; Harlos, K.; Grimes, J.M.; Stuart, D.I. Structure of a pestivirus envelope glycoprotein E2 clarifies its role in cell entry. Cell Rep. 2013, 3, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, J.; Kanai, R.; Modis, Y. Crystal structure of glycoprotein E2 from bovine viral diarrhea virus. Proc. Natl. Acad. Sci. 2013, 110, 6805–6810. [Google Scholar] [CrossRef] [PubMed]
- Iourin, O.; Harlos, K.; El Omari, K.; Lu, W.; Kadlec, J.; Iqbal, M.; Meier, C.; Palmer, A.; Jones, I.; Thomas, C.; et al. Expression, purification and crystallization of the ectodomain of the envelope glycoproteinE2 from Bovine viral diarrhoea virus. Acta. Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Langedijk, J.; van Veelen, P.; Schaaper, W.; de Ru, A.; Meloen, R.; Hulst, M. A structural model of pestivirus Erns based on disulfide bond connectivity and homology modeling reveals an extremely rare vicinal disulfide. J. Virol. 2002, 76, 10383–10392. [Google Scholar] [CrossRef] [PubMed]
- Tews, B.A.; Schürmann, E.M.; Meyers, G. Mutation of cysteine 171 of pestivirus Erns RNase prevents homodimer formation and leads to attenuation of classical swine fever virus. J. Virol. 2009, 83, 4823–4834. [Google Scholar] [CrossRef] [PubMed]
- Van Gennip, H.G.; Hesselink, A.T.; Moormann, R.J.; Hulst, M.M. Dimerization of glycoprotein Erns of classical swine fever virus is not essential for viral replication and infection. Arch. Virol. 2005, 150, 2271–2786. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Sainz, I.; Holinka, L.G.; Gladue, D.; O’Donnell, V.; Lu, Z.; Gavrilov, B.K.; Risatti, G.R.; Borca, M.V. Substitution of specific cysteine residues in the E1 glycoprotein of classical swine fever virus strain Brescia affects formation of E1-E2 heterodimers and alters virulence in swine. J. Virol. 2011, 85, 7264–7272. [Google Scholar] [CrossRef] [PubMed]
- Ronecker, S.; Zimmer, G.; Herrler, G.; Greiser-Wilke, I.; Grummer, B. Formation of bovine viral diarrhea virus E1-E2 heterodimers is essential for virus entry and depends on charged residues in the transmembrane domains. J. Gen. Virol. 2008, 89, 2114–2121. [Google Scholar] [CrossRef] [PubMed]
- Kornfeld, R.; Kornfeld, S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 1985, 54, 631–664. [Google Scholar] [CrossRef] [PubMed]
- Branza-Nichita, N.; Lazar, C.; Dwek, R.A.; Zitzmann, N. Role of N-glycan trimming in the folding and secretion of the pestivirus protein Erns. Biochem. Biophys. Res. Commun. 2004, 319, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Sainz, I.F.; Holinka, L.G.; Lu, Z.; Risatti, G.R.; Borca, M.V. Removal of a N-linked glycosylation site of classical swine fever virus strain Brescia Erns glycoprotein affects virulence in swine. Virology 2008, 370, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Sainz, I.; Holinka, L.G.; Gavrilov, B.K.; Prarat, M.V.; Gladue, D.; Lu, Z.; Jia, W.; Risatti, G.R.; Borca, M.V. Alteration of the N-linked glycosylation condition in E1 glycoprotein of classical swine fever virus strain Brescia alters virulence in swine. Virology 2009, 386, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Risatti, G.; Holinka, L.; Sainz, I.F.; Carrillo, C.; Lu, Z.; Borca, M. N-linked glycosylation status of classical swine fever virus strain Brescia E2 glycoprotein influences virulence in swine. J. Virol. 2007, 81, 924–933. [Google Scholar] [CrossRef] [PubMed]
- Tyborowska, J.; Zdunek, E.; Szewczyk, B. Effect of N-glycosylation inhibition on the synthesis and processing of classical swine fever virus glycoproteins. Acta. Biochim. Pol. 2007, 54, 813–819. [Google Scholar] [PubMed]
- Luo, X.; Pan, R.; Wan, C.; Liu, X.; Wu, J.; Pan, Z. Glycosylation of classical swine fever virus Erns is essential for binding double-stranded RNA and preventing interferon-beta induction. Virus Res. 2009, 146, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Gavrilov, B.K.; Rogers, K.; Fernandez-Sainz, I.J.; Holinka, L.G.; Borca, M.V.; Risatti, G.R. Effects of glycosylation on antigenicity and immunogenicity of classical swine fever virus envelope proteins. Virology 2011, 420, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Langedijk, J.P.M.; Middel, W.G.J.; Meloen, R.H.; Kramps, J.A.; de Smit, J.A. Enzyme-linked immunosorbent assay using a virus type-specific peptide based on a subdomain of envelope protein Erns for serologic diagnosis of pestivirus infections in swine. J. Clin. Microbiol. 2001, 39, 906–912. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Trottier, E.; Pasick, J.; Sabara, M. Identification of antigenic regions of the Erns protein for pig antibodies elicited during classical swine fever virus infection. J. Biochem. 2004, 136, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; McRae, H.; Dan, H.; Tangorra, E.; Laverdiere, A.; Pasick, J. High-resolution epitope mapping for monoclonal antibodies to the structural protein Erns of classical swine fever virus using peptide array and random peptide phage display approaches. J. Gen. Virol. 2010, 91, 2928–2940. [Google Scholar] [CrossRef] [PubMed]
- Christmann, A.; Wentzel, A.; Meyer, C.; Meyers, G.; Kolmar, H. Epitope mapping and affinity purification of monospecific antibodies by Escherichia coli cell surface display of gene-derived random peptide libraries. J. Immunol. Methods 2001, 257, 163–173. [Google Scholar] [CrossRef]
- Zhang, F.; Yu, M.; Weiland, E.; Morrissy, C.J.; Zhang, N.; Westbury, H.A.; Wang, L.F. Characterization of epitopes for neutralizing monoclonal antibodies to classical swine fever virus E2 and Erns using phage-displayed random peptide library. Arch. Virol. 2006, 151, 37–54. [Google Scholar] [CrossRef] [PubMed]
- Meyer, D.; Aebischer, A.; Müller, M.; Grummer, B.; Greiser-Wilke, I.; Moennig, V.; Hofmann, M.A. New insights into the antigenic structure of the glycoprotein Erns of classical swine fever virus by epitope mapping. Virology 2012, 433, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Van Rijn, P.A.; van Gennip, H.G.P.; de Meijer, E.J.; Moormann, R.J.M. A preliminary map of epitopes on envelope glycoprotein El of HCV strain Brescia. Vet. Microbiol. 1992, 33, 221–230. [Google Scholar] [CrossRef]
- Van Rijn, P.A.; van Gennip, H.G.P.; de Meijer, E.J.; Moormann, R.J.M. Epitope mapping of envelope glycoprotein El of hog cholera virus strain Brescia. J. Gen. Virol. 1993, 74, 2053–2060. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.Y.; Huang, C.C.; Deng, M.C.; Huang, Y.L.; Lin, Y.J.; Liu, H.M.; Lin, Y.L.; Wang, F.I. Antigenic mimicking with cysteine-based cyclized peptides reveals a previously unknown antigenic determinant on E2 glycoprotein of classical swine fever virus. Virus Res. 2012, 163, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.Y.; Huang, C.C.; Lin, Y.J.; Deng, M.C.; Tsai, C.H.; Chang, W.M.; Wang, F.I. Identification of antigen-specific residues on E2 glycoprotein of classical swine fever virus. Virus Res. 2010, 152, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Van Rijn, P.A. A common neutralizing epitope on envelope glycoprotein E2 of different pestiviruses: Implications for improvement of vaccines and diagnostics for classical swine fever (CSF)? Vet. Microbiol. 2007, 125, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.P.; Hou, Q.; Xia, Z.H.; Chen, D.; Li, N.; Sun, Y.; Qiu, H.J. Identification of a conserved linear B-cell epitope at the N-terminus of the E2 glycoprotein of classical swine fever virus by phage-displayed random peptide library. Virus Res. 2008, 135, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.; Chen, N.; Liao, X.; Xie, W.; Li, D.; Li, X.; Fang, W. The epitope recognized by monoclonal antibody 2B6 in the B/C domains of classical swine fever virus glycoprotein E2 affects binding to hyperimmune sera and virus replication. J. Microbiol. Biotechnol. 2015, 25, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Lin, F.; Mallory, M.; Clavijo, A. Deletions of structural glycoprotein E2 of classical swine fever virus strain Alfort/187 resolve a linear epitope of monoclonal antibody WH303 and the minimal N-terminal domain essential for binding immunoglobulin G antibodies of a pig hyperimmune serum. J. Virol. 2000, 74, 11619–11625. [Google Scholar] [CrossRef] [PubMed]
- Kortekaas, J.; Vloet, R.P.M.; Weerdmeester, K.; Ketelaar, J.; van Eijk, M.; Loeffen, W.L. Rational design of a classical swine fever C-strain vaccine virus that enables the differentiation between infected and vaccinated animals. J. Virol. Methods 2010, 163, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Wang, L.F.; Shiell, B.J.; Morrissy, C.J.; Westbury, H.A. Fine mapping of a C-terminal linear epitope highly conserved among the major envelope glycoprotein E2 (gp51 to gp54) of different pestiviruses. Virology 1996, 222, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Li, G.X.; Zhou, Y.J.; Yu, H.; Li, L.; Wang, Y.X.; Tong, W.; Hou, J.W.; Xu, Y.Z.; Zhu, J.P.; Xu, A.T.; et al. A novel dendrimeric peptide induces high level neutralizing antibodies against classical swine fever virus in rabbits. Vet. Microbiol. 2012, 156, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Tu, C.; Wang, C.; Yu, X.; Wu, J.; Guo, S.; Shao, M.; Gong, Q.; Zhu, Q.; Kong, X. The protective immune response induced by B cell epitope of classical swine fever virus glycoprotein E2. J. Virol. Methods 2006, 134, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yu, X.; Wang, C.; Wu, J.; Kong, X.; Tu, C. Quadruple antigenic epitope peptide producing immune protection against classical swine fever virus. Vaccine 2006, 24, 7175–7180. [Google Scholar] [CrossRef] [PubMed]
- Monsó, M.; Tarradas, J.; de la Torre, B.G.; Sobrino, F.; Ganges, L.; Andreu, D. Peptide vaccine candidates against classical swine fever virus: T cell and neutralizing antibody responses of dendrimers displaying E2 and NS2–3 epitopes. J. Pept. Sci. 2011, 17, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Reimann, I.; Depner, K.; Utke, K.; Leifer, I.; Lange, E.; Beer, M. Characterization of a new chimeric marker vaccine candidate with a mutated antigenic E2-epitope. Vet. Microbiol. 2010, 142, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Tarradas, J.; Monsó, M.; Muñoz, M.; Rosell, R.; Fraile, L.; Frías, M.T.; Domingo, M.; Andreu, D.; Sobrino, F.; Ganges, L. Partial protection against classical swine fever virus elicited by dendrimeric vaccine-candidate peptides in domestic pigs. Vaccine 2011, 29, 4422–4429. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Zhang, B.Q.; Shen, Z.; Chen, Y.H. Antigens containing TAVSPTTLR tandem repeats could be used in assaying antibodies to classical swine fever virus. Acta Virol. 2009, 53, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Paton, D.J.; Lowings, J.P.; Barrett, A.D. Epitope mapping of the gp53 envelope protein of bovine viral diarrhea virus. Virology 1992, 190, 763–772. [Google Scholar] [CrossRef]
- Deregt, D.; van Rijn, P.A.; Wiens, T.Y.; van den Hurk, J. Monoclonal antibodies to the E2 protein of a new genotype (type 2) of bovine viral diarrhea virus define three antigenic domains involved in neutralization. Virus Res. 1998, 57, 171–181. [Google Scholar] [CrossRef]
- Jelsma, H.; Loeffen, W.L.; van Beuningen, A.; van Rijn, P.A. Preliminary mapping of non-conserved epitopes on envelope glycoprotein E2 of Bovine viral diarrhea virus type 1 and 2. Vet. Microbiol. 2013, 166, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Deregt, D.; Bolin, S.R.; van den Hurk, J.; Ridpath, J.F.; Gilbert, S.A. Mapping of a type 1-specific and a type-common epitope on the E2 (gp53) protein of bovine viral diarrhea virus with neutralization escape mutants. Virus Res. 1998, 53, 81–90. [Google Scholar] [CrossRef]
- Hulst, M.M.; Moormann, R.J. Inhibition of pestivirus infection in cell culture by envelope proteins Erns and E2 of classical swine fever virus: Erns and E2 interact with different receptors. J. Gen. Virol. 1997, 78, 2779–2787. [Google Scholar] [PubMed]
- Wang, Z.; Nie, Y.; Wang, P.; Ding, M.; Deng, H. Characterization of classical swine fever virus entry by using pseudotyped viruses: E1 and E2 are sufficient to mediate viral entry. Virology 2004, 330, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.L.; Sainz, I.F.; Ansari, I.H.; Gil, L.H.; Vassilev, V.; Donis, R.O. The envelope glycoprotein E2 is a determinant of cell culture tropism in ruminant pestiviruses. J. Gen. Virol. 2003, 84, 1269–1274. [Google Scholar] [CrossRef] [PubMed]
- Reimann, I.; Depner, K.; Trapp, S.; Beer, M. An avirulent chimeric Pestivirus with altered cell tropism protects pigs against lethal infection with classical swine fever virus. Virology 2004, 322, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Asfor, A.S.; Wakeley, P.R.; Drew, T.W.; Paton, D.J. Recombinant pestivirus E2 glycoproteins prevent viral attachment to permissive and non permissive cells with different efficiency. Virus Res. 2014, 189, 147–57. [Google Scholar] [CrossRef] [PubMed]
- Hulst, M.M.; van Gennip, H.G.; Vlot, A.C.; Schooten, E.; de Smit, A.J.; Moormann, R.J. Interaction of classical swine fever virus with membrane-associated heparan sulfate: Role for virus replication in vivo and virulence. J. Virol. 2001, 75, 9585–9595. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.; Flick-Smith, H.; McCauley, J.W. Interactions of bovine viral diarrhoea virus glycoprotein Erns with cell surface glycosaminoglycans. J. Gen. Virol. 2000, 81, 451–459. [Google Scholar] [PubMed]
- Chen, J.; He, W.R.; Shen, L.; Dong, H.; Yu, J.; Wang, X.; Yu, S.; Li, Y.; Li, S.; Luo, Y.; et al. The laminin receptor is an attachment cellular receptor for classical swine fever virus. J. Virol. 2015, 89, 4894–4906. [Google Scholar] [CrossRef] [PubMed]
- Maurer, K.; Krey, T.; Moennig, V.; Thiel, H.J.; Rümenapf, T. CD46 is a cellular receptor for bovine viral diarrhea virus. J. Virol. 2004, 78, 1792–1799. [Google Scholar] [CrossRef] [PubMed]
- Dräger, C.; Beer, M.; Blome, S. Porcine complement regulatory protein CD46 and heparan sulfates are the major factors for classical swine fever virus attachment in vitro. Arch. Virol. 2015, 160, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Krey, T.; Himmelreich, A.; Heimann, M.; Menge, C.; Thiel, H.J.; Maurer, K.; Rümenapf, T. Function of bovine CD46 as a cellular receptor for bovine viral diarrhea virus is determined by complement control protein 1. J. Virol. 2006, 80, 3912–3922. [Google Scholar] [CrossRef] [PubMed]
- Zezafoun, H.; Decreux, A.; Desmecht, D. Genetic and splice variations of Bostaurus CD46 shift cell permissivity to BVDV, the bovine pestivirus. Vet. Microbiol. 2011, 152, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, L.; Zhao, D.; Zhang, G.; Luo, J.; Deng, R.; Yang, Y. Identification of host cell binding peptide from an overlapping peptide library for inhibition of classical swine fever virus infection. Virus Genes 2011, 43, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Schelp, C.; Greiser-Wilke, I.; Moennig, V. An actin-binding protein is involved in pestivirus entry into bovine cells. Virus Res. 2000, 68, 1–5. [Google Scholar] [CrossRef]
- He, F.; Ling, L.; Liao, Y.; Li, S.; Han, W.; Zhao, B.; Sun, Y.; Qiu, H.J. Beta-actin interacts with the E2 protein and is involved in the early replication of classical swine fever virus. Virus Res. 2014, 179, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Shi, Z.; Guo, H.; Qu, H.; Zhang, Y.; Tu, C. Annexin 2 is a host protein binding to classical swine fever virus E2 glycoprotein and promoting viral growth in PK-15 cells. Virus Res. 2015, 201, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Gladue, D.P.; Baker-Bransetter, R.; Holinka, L.G.; Fernandez-Sainz, I.J.; O’Donnell, V.; Fletcher, P.; Lu, Z.; Borca, M.V. Interaction of CSFV E2 protein with swine host factors as detected by yeast two-hybrid system. PLoS ONE 2014, 9, e85324. [Google Scholar] [CrossRef] [PubMed]
- Garry, R.F.; Dash, S. Proteomics computational analyses suggest that hepatitis C virus E1 and pestivirus E2 envelope glycoproteins are truncated class II fusion proteins. Virology 2003, 307, 255–265. [Google Scholar] [CrossRef]
- Fernández-Sainz, I.; Largo, E.; Gladue, D.; Fletcher, P.; O’Donnell, V.; Holinka, L.; Carey, L.; Lu, X.; Nieva, J.; Borca, M. Effect of specific amino acid substitutions in the putative fusion peptide of structural glycoprotein E2 on classical swine fever virus replication. Virology 2014, 456, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Grummer, B.; Grotha, S.; Greiser-Wilke, I. Bovine viral diarrhoea virus is internalized by clathrin-dependent receptor mediated endocytosis. J. Vet. Med. B Infect. Dis. Vet. Public Health 2004, 51, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Lecot, S.; Belouzard, S.; Dubuisson, J.; Rouillé, Y. Bovine viral diarrhea virus entry is dependent on clathrin-mediated endocytosis. J. Virol. 2005, 79, 10826–10829. [Google Scholar] [CrossRef] [PubMed]
- Krey, T.; Thiel, H.J.; Rümenapf, T. Acid-resistant bovine pestivirus requires activation for pH-triggered fusion during entry. J. Virol. 2005, 79, 4191–4200. [Google Scholar] [CrossRef] [PubMed]
- Kampmann, T.; Mueller, D.S.; Mark, A.E.; Young, P.R.; Kobe, B. The Role of histidine residues in low-pH-mediated viral membrane fusion. Structure 2006, 14, 1481–1487. [Google Scholar] [CrossRef] [PubMed]
- Zürcher, C.; Sauter, K.S.; Mathys, V.; Wyss, F.; Schweizer, M. Prolonged activity of the pestiviral RNase Erns as an interferon antagonist after uptake by clathrin-mediated endocytosis. J. Virol. 2014, 88, 7235–7243. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Shi, H.; Ren, Y.; Guo, F.; Ni, W.; Qiao, J.; Wang, P.; Zhang, H.; Chen, C. Bovine viral diarrhea virus infection induces autophagy in MDBK cells. J. Microbiol. 2014, 52, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Zhao, M.; Ye, Z.; Gou, H.; Wang, J.; Yi, L.; Dong, X.; Liu, W.; Luo, Y.; Liao, M.; et al. Autophagy enhances the replication of classical swine fever virus in vitro. Autophagy 2014, 10, 93–110. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Shi, H.; Shi, M.; Meng, L.; Bao, H.; Zhang, G.; Ren, Y.; Zhang, H.; Guo, F.; Qiao, J.; et al. Roles of bovine viral diarrhea virus envelope glycoproteins in inducing autophagy in MDBK cells. Microb. Pathog. 2014, 76, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Branza-Nichita, N.; Durantel, D.; Carrouée-Durantel, S.; Dwek, R.A.; Zitzmann, N. Antiviral effect of N-butyldeoxynojirimycin against bovine viral diarrhea virus correlates with misfolding of E2 envelope proteins and impairment of their association into E1-E2 heterodimers. J. Virol. 2001, 75, 3527–3536. [Google Scholar] [CrossRef] [PubMed]
- Armengol, E.; Wiesmüller, K.H.; Wienhold, D.; Büttner, M.; Pfaff, E.; Jung, G.; Saalmüller, A. Identification of T-cell epitopes in the structural and non-structural proteins of classical swine fever virus. J. Gen. Virol. 2002, 83, 551–560. [Google Scholar] [PubMed]
- Ceppi, M.; de Bruin, M.G.; Seuberlich, T.; Balmelli, C.; Pascolo, S.; Ruggli, N.; Wienhold, D.; Tratschin, J.D.; McCullough, K.C.; Summerfield, A. Identification of classical swine fever virus protein E2 as a target for cytotoxic T cells by using mRNA-transfected antigen-presenting cells. J. Gen. Virol. 2005, 86, 2525–2534. [Google Scholar] [CrossRef] [PubMed]
- Franzoni, G.; Kurkure, N.V.; Essler, S.E.; Pedrera, M.; Everett, H.E.; Bodman-Smith, K.B.; Crooke, H.R.; Graham, S.P. Proteome-wide screening reveals immunodominance in the CD8 T cell response against classical swine fever virus with antigen-specificity dependent on MHC class I haplotype expression. PLoS ONE 2013, 8, e84246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruschke, C.J.; Hulst, M.M.; Moormann, R.J.; van Rijn, P.A.; van Oirschot, J.T. Glycoprotein Erns of pestiviruses induces apoptosis in lymphocytes of several species. J. Virol. 1997, 71, 6692–6696. [Google Scholar] [PubMed]
- Magkouras, I.; Mätzener, P.; Rümenapf, T.; Peterhans, E.; Schweizer, M. RNase-dependent inhibition of extracellular, but not intracellular, dsRNA-induced interferon synthesis by Erns of pestiviruses. J. Gen. Virol. 2008, 89, 2501–2506. [Google Scholar] [CrossRef] [PubMed]
- Mätzener, P.; Magkouras, I.; Rümenapf, T.; Peterhans, E.; Schweizer, M. The viral RNase Erns prevents IFN type-I triggering by pestiviral single- and double-stranded RNAs. Virus Res. 2009, 140, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Meyers, G.; Ege, A.; Fetzer, C.; von Freyburg, M.; Elbers, K.; Carr, V.; Prentice, H.; Charleston, B.; Schürmann, E.M. Bovine viral diarrhea virus: Prevention of persistent fetal infection by a combination of two mutations affecting Erns RNase and Npro protease. J. Virol. 2007, 81, 3327–3338. [Google Scholar] [CrossRef] [PubMed]
- Python, S.; Gerber, M.; Suter, R.; Ruggli, N.; Summerfield, A. Efficient sensing of infected cells in absence of virus particles by plasmacytoid dendritic cells is blocked by the viral ribonuclease Erns. PLoS Pathog. 2013, 9, e1003412. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Pan, Z.; Zhang, C. The selection pressure analysis of classical swine fever virus envelope protein genes Erns and E2. Virus Res. 2008, 131, 132–135. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Pei, J.; Bai, J.; Zhao, M.; Ju, C.; Yi, L.; Kang, Y.; Zhang, X.; Chen, L.; Li, Y.; et al. Genetic diversity and positive selection analysis of classical swine fever virus isolates in south China. Virus Genes 2011, 43, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Pérez, L.J.; Díaz de Arce, H.; Perera, C.L.; Rosell, R.; Frías, M.T.; Percedo, M.I.; Tarradas, J.; Dominguez, P.; Núñez, J.I.; Ganges, L. Positive selection pressure on the B/C domains of the E2-gene of classical swine fever virus in endemic areas under C-strain vaccination. Infect. Genet. Evol. 2012, 12, 1405–1412. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Zhang, C. Evidence for positive selection on the E2 gene of bovine viral diarrhoea virus type 1. Virus Genes 2007, 35, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Leifer, I.; Ruggli, N.; Blome, S. Approaches to define the viral genetic basis of classical swine fever virus virulence. Virology 2013, 438, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.; Guo, Z.; Ding, N.; He, C. Studying classical swine fever virus: Making the best of a bad virus. Virus Res. 2015, 197, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.; von Freyburg, M.; Elbers, K.; Meyers, G. Recovery of virulent and RNase-negative attenuated type 2 bovine viral diarrhea viruses from infectious cDNA clones. J. Virol. 2002, 76, 8494–8503. [Google Scholar] [CrossRef] [PubMed]
- Risatti, G.; Holinka, L.; Lu, Z.; Kutish, G.; Tulman, E.; French, R.; Sur, J.; Rock, D.; Borca, M. Mutation of E1 glycoprotein of classical swine fever virus affects viral virulence in swine. Virology 2005, 343, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Risatti, G.; Borca, M.; Kutish, G.; Lu, Z.; Holinka, L.; French, R.; Tulman, E.; Rock, D. The E2 glycoprotein of classical swine fever virus is a virulence determinant in swine. J. Virol. 2005, 79, 3787–3796. [Google Scholar] [CrossRef] [PubMed]
- Risatti, G.; Holinka, L.; Fernandez Sainz, I.; Carrillo, C.; Kutish, G.; Lu, Z.; Zhu, J.; Rock, D.; Borca, M. Mutations in the carboxyl terminal region of E2 glycoprotein of classical swine fever virus are responsible for viral attenuation in swine. Virology 2007, 364, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Risatti, G.; Holinka, L.; Carrillo, C.; Kutish, G.; Lu, Z.; Tulman, E.; Sainz, I.F.; Borca, M. Identification of a novel virulence determinant within the E2 structural glycoprotein of classical swine fever virus. Virology 2006, 355, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Sakoda, Y.; Yoshino, F.; Nomura, T.; Yamamoto, N.; Sato, Y.; Okamatsu, M.; Ruggli, N.; Kida, H. Selection of classical swine fever virus with enhanced pathogenicity reveals synergistic virulence determinants in E2 and NS4B. J. Virol. 2012, 86, 8602–8613. [Google Scholar] [CrossRef] [PubMed]
- Fahnøe, U.; Pedersen, A.G.; Risager, P.C.; Nielsen, J.; Belsham, G.J.; Höper, D.; Beer, M.; Rasmussen, T.B. Rescue of the highly virulent classical swine fever virus strain “Koslov” from cloned cDNA and first insights into genome variations relevant for virulence. Virology 2014, 468–470, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Van Gennip, H.G.; Vlot, A.C.; Hulst, M.M.; de Smit, A.J.; Moormann, R.J. Determinants of virulence of classical swine fever virus strain Brescia. J. Virol. 2004, 78, 8812–8823. [Google Scholar] [CrossRef] [PubMed]
- Branza-Nichita, N.; Lazar, C.; Durantel, D.; Dwek, R.A.; Zitzmann, N. Role of disulfide bond formation in the folding and assembly of the envelope glycoproteins of a pestivirus. Biochem. Biophys. Res. Commun. 2002, 296, 470–476. [Google Scholar] [CrossRef]
- Aebischer, A.; Müller, M.; Hofmann, M.A. Two newly developed Erns-based ELISAs allow the differentiation of Classical Swine Fever virus-infected from marker-vaccinated animals and the discrimination of pestivirus antibodies. Vet. Microbiol. 2013, 161, 274–285. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.Y.; Wu, C.W.; Lin, G.J.; Lee, W.C.; Chien, M.S.; Huang, C. Enhancing expression of the classical swine fever virus glycoprotein E2 in yeast and its application to a blocking ELISA. J. Biotechnol. 2014, 174, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, S.; von Rosen, T.; Blome, S.; Loeffen, W.; Haegeman, A.; Koenen, F.; Uttenthal, Å. Evaluation of classical swine fever virus antibody detection assays with an emphasis on the differentiation of infected from vaccinated animals. Rev. Sci. Tech. Off. Int. Epiz. 2012, 31, 997–1010. [Google Scholar]
- Windisch, J.M.; Schneider, R.; Stark, R.; Weiland, E.; Meyers, G.; Thiel, H.J. RNase of classical swine fever virus: Biochemical characterization and inhibition by virus-neutralizing monoclonal antibodies. J. Virol. 1996, 70, 352–358. [Google Scholar] [PubMed]
- Chen, N.; Hu, H.; Zhang, Z.; Shuai, J.; Jiang, L.; Fang, W. Genetic diversity of the envelope glycoprotein E2 of classical swine fever virus: Recent isolates branched away from historical and vaccine strains. Vet. Microbiol. 2008, 127, 286–299. [Google Scholar] [CrossRef] [PubMed]
- Bouma, A.; de Smit, A.J.; de Kluijver, E.P.; Terpstra, C.; Moormann, R.J.M. Efficacy and stability of a subunit vaccine based on glycoprotein E2 of classical swine fever virus. Vet. Microbiol. 1999, 66, 101–114. [Google Scholar] [CrossRef]
- Bouma, A.; de Smit, A.J.; de Jong, M.C.M.; de Kluijver, E.P.; Moormann, R.J.M. Determination of the onset of the herd-immunity induced by the E2 sub-unit vaccine against classical swine fever virus. Vaccine 2000, 18, 1374–1381. [Google Scholar] [CrossRef]
- Hulst, M.M.; Westra, D.F.; Wensvoort, G.; Moormann, R.J. Glycoprotein E1 of hog cholera virus expressed in insect cells protects swine from hog cholera. J. Virol. 1993, 67, 5435–5442. [Google Scholar] [PubMed]
- Rümenapf, T.; Stark, R.; Meyers, G.; Thiel, H.J. Structural proteins of hog cholera virus expressed by vaccinia virus: Further characterization and induction of protective immunity. J. Virol. 1991, 65, 589–597. [Google Scholar] [PubMed]
- Van Zijl, M.; Wensvoort, G.; de Kluyver, E.; Hulst, M.; van der Gulden, H.; Gielkens, A.; Berns, A.; Moormann, R. Live attenuated pseudorabies virus expressing envelope glycoprotein E1 of hog cholera virus protects swine against both pseudorabies and hog cholera. J. Virol. 1991, 65, 2761–2765. [Google Scholar] [PubMed]
- Uttenthal, Å.; le Potier, M.F.; Romero, L.; de Mia, G.M.; Floegel-Niesmann, G. Classical swine fever (CSF) marker vaccine-Trial I. Challenge studies in weaner pigs. Vet. Microbiol. 2001, 83, 85–106. [Google Scholar] [CrossRef]
- Van Oirschot, J.T. Vaccinology of classical swine fever: From lab to field. Vet. Microbiol. 2003, 96, 367–384. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Tong, C.; Li, D.; Wan, J.; Yuan, X.; Li, X.; Peng, J.; Fang, W. Antigenic analysis of classical swine fever virus E2 glycoprotein using pig antibodies identifies residues contributing to antigenic variation of the vaccine C-strain and group 2 strains circulating in China. Virol. J. 2010, 7. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Nishi, K.; Macleod, E.; Sabara, M.I.; Lin, M.; Handel, K.; Pasick, J. Baculovirus expression and antigenic characterization of classical swine fever virus E2 proteins. Transbound. Emerg. Dis. 2013, 60, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.; Niu, D.D.; Si, H.L.; Ding, N.Z.; He, C.Q. Vaccination influences the evolution of classical swine fever virus. Infect. Genet. Evol. 2014, 25, 69–77. [Google Scholar] [CrossRef] [PubMed]
- De Arce, H.D.; Ganges, L.; Barrera, M.; Naranjo, D.; Sobrino, F.; Frías, M.T.; Núñez, J.I. Origin and evolution of viruses causing classical swine fever in Cuba. Virus Res. 2005, 112, 123–131. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.-I.; Deng, M.-C.; Huang, Y.-L.; Chang, C.-Y. Structures and Functions of Pestivirus Glycoproteins: Not Simply Surface Matters. Viruses 2015, 7, 3506-3529. https://doi.org/10.3390/v7072783
Wang F-I, Deng M-C, Huang Y-L, Chang C-Y. Structures and Functions of Pestivirus Glycoproteins: Not Simply Surface Matters. Viruses. 2015; 7(7):3506-3529. https://doi.org/10.3390/v7072783
Chicago/Turabian StyleWang, Fun-In, Ming-Chung Deng, Yu-Liang Huang, and Chia-Yi Chang. 2015. "Structures and Functions of Pestivirus Glycoproteins: Not Simply Surface Matters" Viruses 7, no. 7: 3506-3529. https://doi.org/10.3390/v7072783






