Development and Validation of a Multiplex Reverse Transcription PCR Assay for Simultaneous Detection of Three Papaya Viruses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Viruses
2.2. Primer Design
| Virus | Primer a | Sequence (5’-3’) | Size (bp) | Target Gene | Location (nt) b |
|---|---|---|---|---|---|
| PRSV | PRSV953-F | GCGATGCTCATAAACATACCTGA | 953 | HC-Pro/P3 | 2774–2796 |
| PRSV953-R | TGTACACAGTACTTCGGTGAGAAGTCGTA | 3698–3726 | |||
| PRSV613-F | TTGTGTACGACTTCTCACCGAA | 613 | P3 | 3693–3714 | |
| PRSV613-R | CGAATGTCATCCAAAGACTGATGATAAAC | 4277–4305 | |||
| PRSV515-F | TTGTGTACGACTTCTCACCGAA | 515 | P3 | 3693–3714 | |
| PRSV515-R | CACATCAATCATCATCAAAATTAATGT | 4181–4207 | |||
| PapMV | PapMV476-F | ATGGTAGCTGCTAAGGTTCCAGC | 476 | CP | 6024–6046 |
| PapMV476-R | GACCCAGAAATTTGGCCTTTGGTGATG | 6473–6499 | |||
| PapMV205-F | CCAAATTTGCCGCGTTCGACT | 205 | CP | 6295–6315 | |
| PapMV205-R | GACCCAGAAATTTGGCCTTTGGTGATG | 6473–6499 | |||
| PLDMV | PLDMV355-F | GGCATGTGGTTTATGATGCAAGGG | 355 | CP | 9528–9551 |
| PLDMV355-R | GCTCCGTGTTCTCAGTCGCATT | 9861–9882 |
2.3. Nucleic Acid Extraction
2.4. Reverse Transcription
2.5. The Uniplex PCR
2.6. Optimization of the Multiplex PCR Conditions
2.7. The Sensitivity of the Uniplex PCR and Multiplex PCR Assays
2.8. The Detection of Viruses in Field Samples by Uniplex RT-PCR and Multiplex RT-PCR
3. Results
3.1. Specificity and Compatibility of Primer Pairs
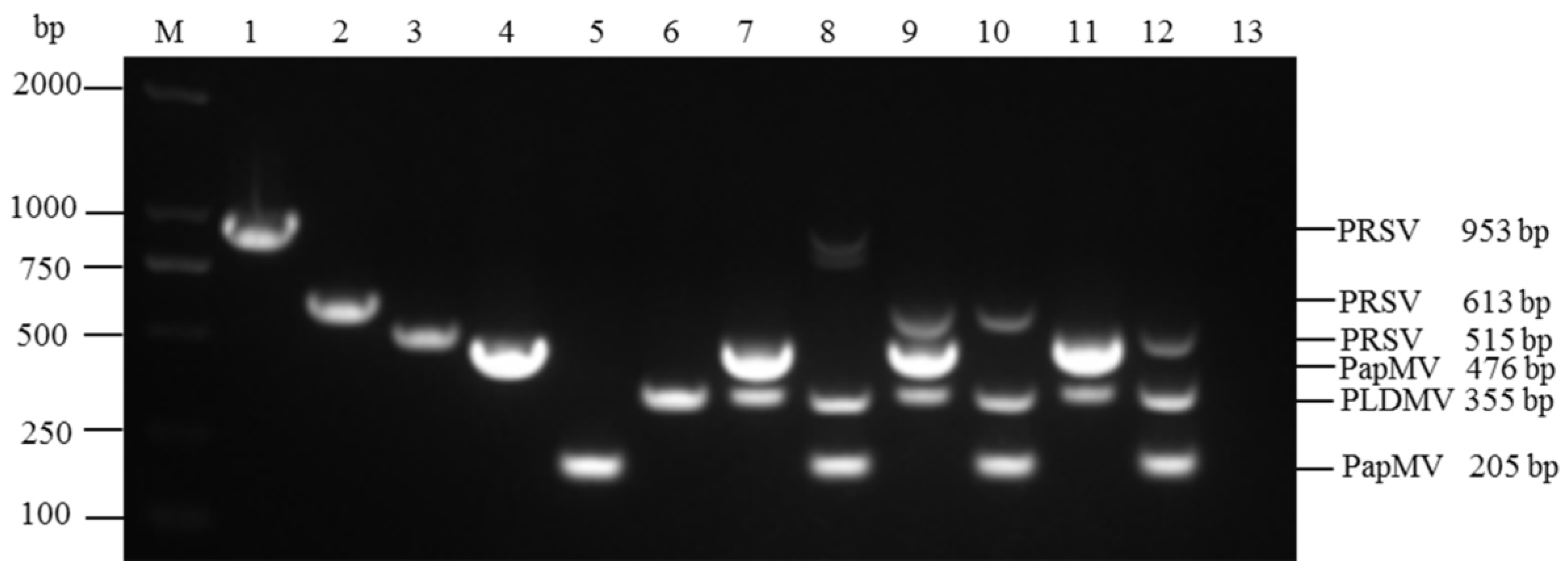
3.2. Optimization of Multiplex RT-PCR
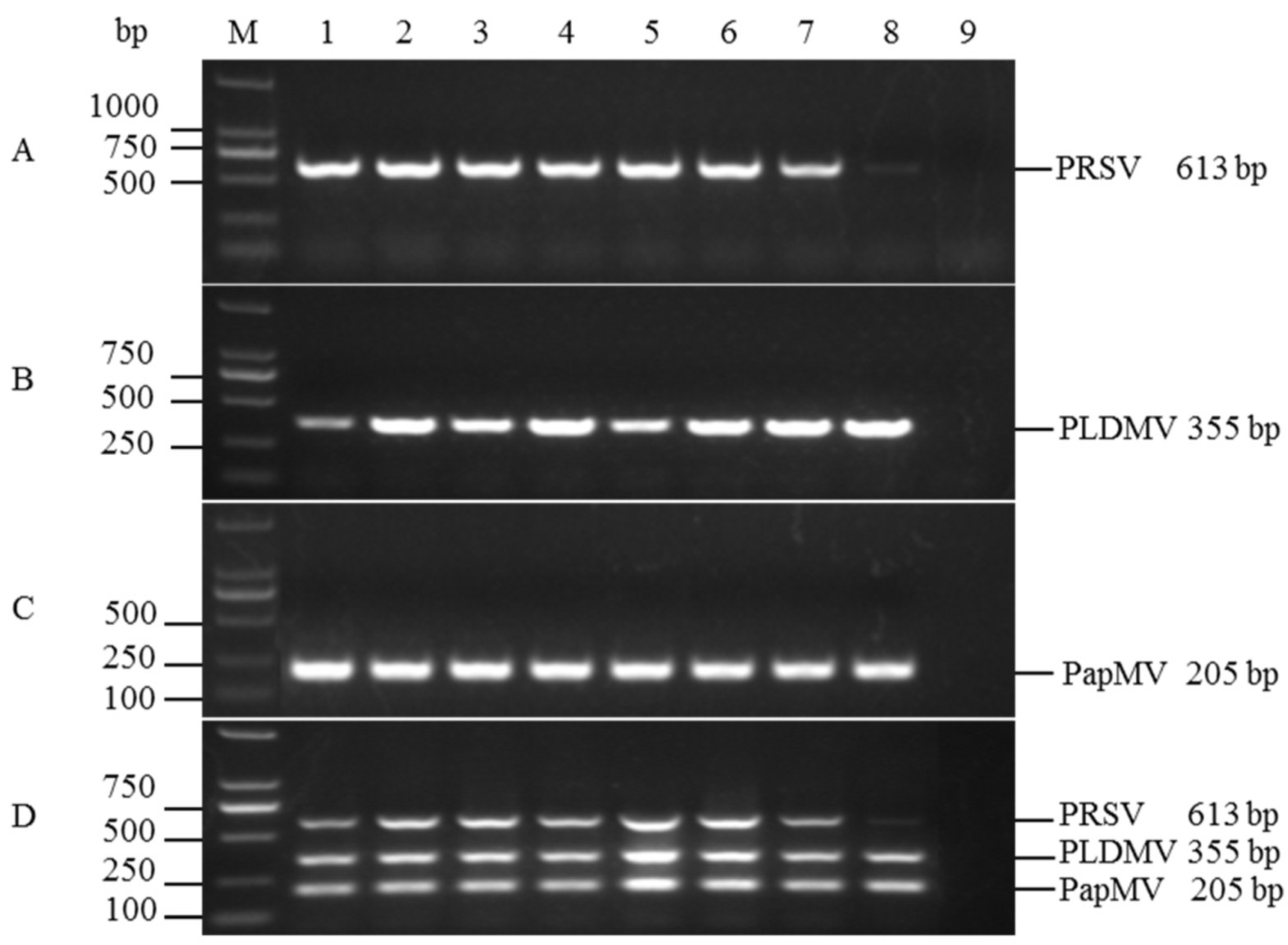

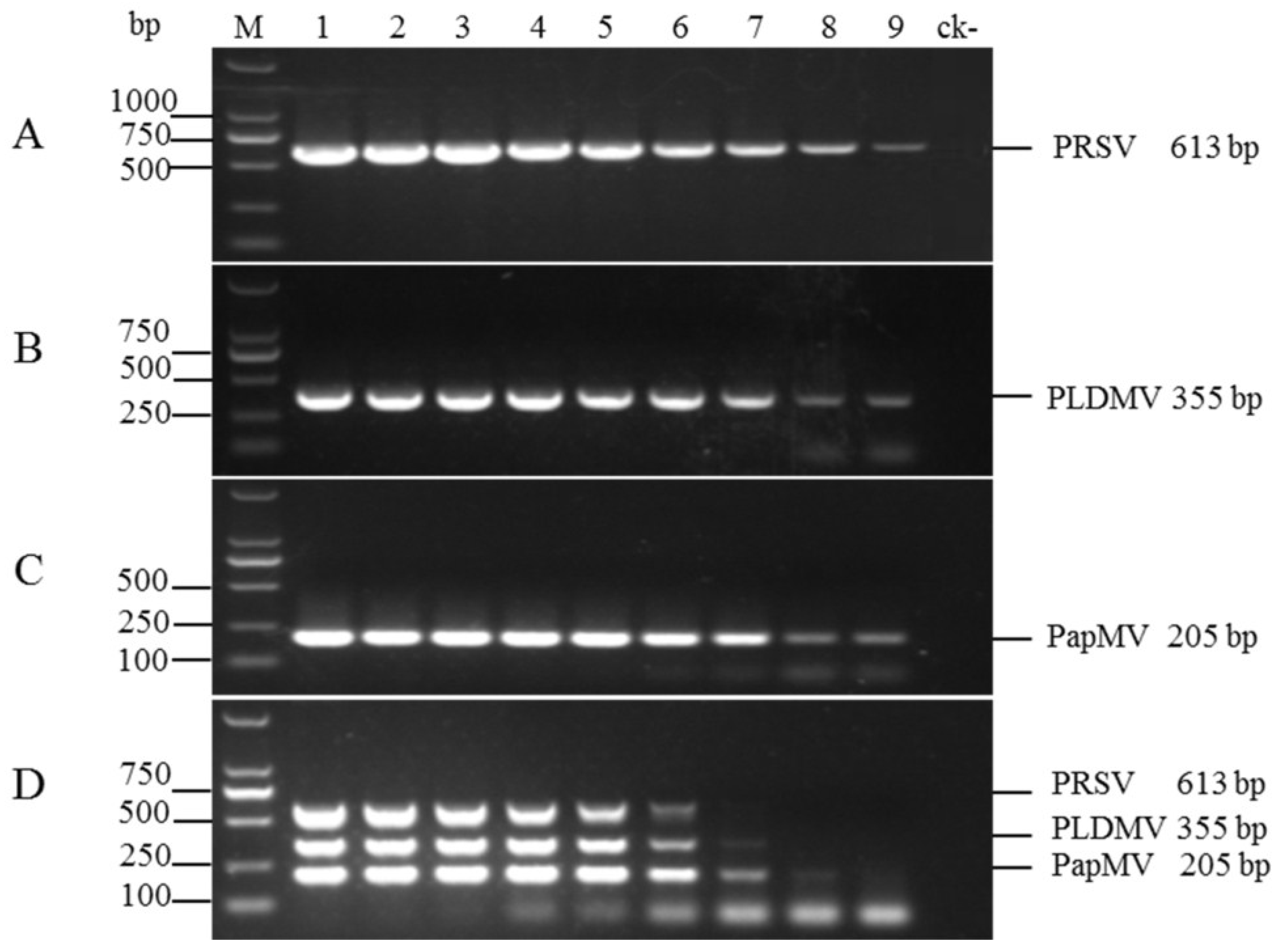
3.3. Sensitivities of the Uniplex and Multiplex RT-PCR
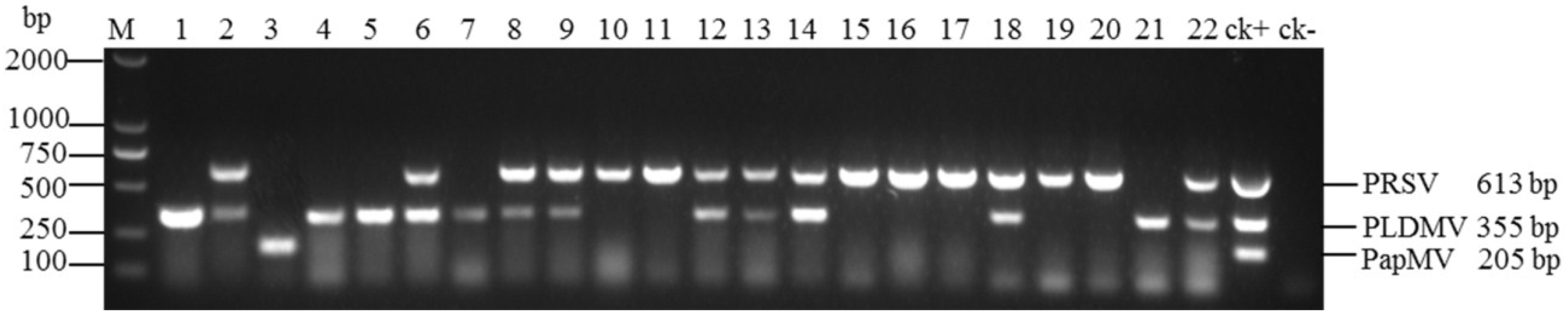
3.4. Evaluation of Multiplex RT-PCR Using Field Samples
| Geographical Region | No. of Samples | PRSV Only | PLDMV Only | PapMV Only | PRSV and PLDMV (Mixed Infection) |
|---|---|---|---|---|---|
| Haikou | 30 | 15(50%) | 8(26.7%) | 1(3.3%) | 6(20%) |
| Sanya | 28 | 17(60.7%) | 8(28.6%) | 0(0%) | 3(10.7%) |
| Wenchang | 16 | 10(62.5%) | 3(18.8%) | 0(0%) | 3(18.8%) |
| Qionghai | 13 | 7(53.8%) | 2(15.4%) | 0(0%) | 4(30.8%) |
| Wanning | 9 | 6(66.7%) | 2(22.2%) | 0(0%) | 1(11.1%) |
| Wuzhishan | 13 | 8(61.5%) | 3(23.1%) | 0(0%) | 2(15.4%) |
| Dongfang | 34 | 9(26.5%) | 13(38.2%) | 1(2.9%) | 11(32.4%) |
| Danzhou | 12 | 8(66.7%) | 3(25%) | 0(0%) | 1(8.3%) |
| Lingao | 15 | 11(73.3%) | 4(26.7%) | 0(0%) | 0(0%) |
| Chengmai | 13 | 6(46.2%) | 4(30.8%) | 0(0%) | 3(23.1%) |
| Dingan | 16 | 16(100%) | 0(0%) | 0(0%) | 0(0%) |
| Tunchang | 14 | 10(71.4%) | 2(14.3%) | 0(0%) | 2(14.3%) |
| Changjiang | 20 | 14(70%) | 4(20%) | 0(0%) | 2(10%) |
| Baisha | 12 | 9(75%) | 1(8.3%) | 0(0%) | 2(16.7%) |
| Qiongzhong | 18 | 8(44.4%) | 8(44.4%) | 0(0%) | 2(11.1%) |
| Lingshui | 19 | 11(57.9%) | 7(36.8%) | 0(0%) | 1(5.3%) |
| Baoting | 21 | 10(47.6%) | 6(28.6%) | 0(0%) | 5(23.8%) |
| Ledong | 38 | 11(28.9%) | 15(39.5%) | 1(2.6%) | 11(28.9%) |
| Total | 341 | 186(54.5%) | 93(27.3%) | 3(0.9%) | 59(17.3%) |
4. Discussion
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References and Notes
- Manshardt, R.M. Papaya. In Biotechnology of Perennial Fruit Crops (Biotechnology in Agraculture No. 8); Hammerschlag, F.A., Litz, R.E., Eds.; CIBA: Wallingford, UK, 1992; pp. 489–511. [Google Scholar]
- Tripathi, S.; Suzuki, J.Y.; Ferreira, S.A.; Gonsalves, D. Papaya ringspot virus-P: Characteristics, pathogenicity, sequence variability and control. Mol. Plant Pathol. 2008, 9, 269–280. [Google Scholar] [CrossRef]
- Maoka, T.; Kashiwazaki, S.; Tsuda, S.; Usugi, T.; Hibino, H. Nucleotide sequence of the capsid protein gene of papaya leaf-distortion mosaic potyvirus. Arch. Virol. 1996, 141, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Ferwerda-Licha, M. Mixed infection of papaya ringspot virus, zucchini yellow mosaic virus and papaya bunchy top affecting papaya (Carica papaya L.) in Puerto Rico. Phytopathology 2002, 92 (Suppl. 6), S25. [Google Scholar]
- Wang, Y.; Shen, W.; Wang, S.; Tuo, D.; Yan, P.; Li, X.; Zhou, P. Complete genomic sequence of Papaya mosaic virus isolate from hainan island, China. Chin. J. Trop. Crops 2013, 34, 297–300. [Google Scholar]
- Chang, L.S.; Lee, Y.S.; Su, H.J.; Hung, T.H. First report of papaya leaf curl virus Infecting papaya plants in Taiwan. Plant Dis. 2003, 87, 204. [Google Scholar] [CrossRef]
- Singh-Pant, P.; Pant, P.; Mukherjee, S.K.; Mazumdar-Leighton, S. Spatial and temporal diversity of begomoviral complexes in papayas with leaf curl disease. Arch. Virol. 2012, 157, 1217–1232. [Google Scholar] [CrossRef] [PubMed]
- Pramesh, D.; Mandal, B.; Phaneendra, C.; Muniyappa, V. Host range and genetic diversity of croton yellow vein mosaic virus, a weed-infecting monopartite begomovirus causing leaf curl disease in tomato. Arch. Virol. 2013, 158, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.-H.; Conover, R.A. Incidence and distribution of papaya viruses in Southern Florida. Plant Dis. 1983, 67, 353–356. [Google Scholar] [CrossRef]
- Hernandez, R.; Suazo, M.; Toledo, P. The papaya apical necrosis virus, a new viral disease in Villa Clara, Cuba. Cienc. Tec. Agric. Prot. Plantas 1990, 13, 29–36. [Google Scholar]
- Gonsalves, D.; Trujillo, E.E. Tomato spotted wilt virus in papaya and detection of the virus by ELISA. Plant Dis. 1986, 70, 501–506. [Google Scholar] [CrossRef]
- Amaral, P.P.; Resende, R.O.; Júnior, M.T.S. Papaya Lethal Yellowing Virus (PLYV) infects vasconcellea cauliflora. Fitopatol. Bras. 2006, 31, 517–517. [Google Scholar] [CrossRef]
- Pereira, A.J.; Alfenas-Zerbini, P.; Cascardo, R.S.; Andrade, E.C.; Murilo Zerbini, F. Analysis of the full-length genome sequence of papaya lethal yellowing virus (PLYV), determined by deep sequencing, confirms its classification in the genus Sobemovirus. Arch. Virol. 2012, 157, 2009–2011. [Google Scholar] [CrossRef] [PubMed]
- Tuo, D.; Shen, W.; Yan, P.; Li, C.; Gao, L.; Li, X.; Li, H.; Zhou, P. Complete genome sequence of an isolate of papaya leaf distortion mosaic virus from commercialized PRSV-resistant transgenic papaya in China. Acta Virol. 2013, 57, 452–455. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.W.; Shen, W.T.; Zhou, P.; Tang, Q.J.; Niu, Y.M.; Peng, M.; Xiong, Z. Complete genomic sequence of a Papaya ringspot virus isolate from Hainan Island, China. Arch. Virol. 2008, 153, 991–993. [Google Scholar] [CrossRef] [PubMed]
- Bau, H.J.; Kung, Y.J.; Raja, J.A.; Chan, S.J.; Chen, K.C.; Chen, Y.K.; Wu, H.W.; Yeh, S.D. Potential threat of a new pathotype of Papaya leaf distortion mosaic virus infecting transgenic papaya resistant to Papaya ringspot virus. Phytopathology 2008, 98, 848–856. [Google Scholar] [CrossRef] [PubMed]
- Cruz, F.C.S.; Tanada, J.M.; Elvira, P.R.V.; Dolores, L.M.; Magdalita, P.M.; Hautea, D.M.; Hautea, R.A. Detection of Mixed Virus Infection with Papaya ringspot virus (PRSV) in Papaya (Carica papaya L.) grown in Luzon, Philippines. Philipp. J. Crop Sci. 2009, 34, 62–74. [Google Scholar]
- Noa-Carrazana, J.C.; González-de-León, D.; Ruiz-Castro, B.S.; Piñero, D.; Silva-Rosales, L. Distribution of Papaya ringspot virus and Papaya mosaic virus in Papaya Plants (Carica papaya) in Mexico. Plant Dis. 2006, 90, 1004–1011. [Google Scholar] [CrossRef]
- Ling, K.; Namba, S.; Gonsalves, C.; Slightom, J.L.; Gonsalves, D. Protection against detrimental effects of potyvirus infection in transgenic tobacco plants expressing the papaya ringspot virus coat protein gene. Biotechnol. N. Y. 1991, 9, 752–758. [Google Scholar] [CrossRef]
- Chiang, C.-H.; Wang, J.-J.; Jan, F.-J.; Yeh, S.-D.; Gonsalves, D. Comparative reactions of recombinant papaya ringspot viruses with chimeric coat protein genes and vild. J. Gen. Virol. 2001, 82, 2827–2836. [Google Scholar] [PubMed]
- Majumder, S.; Baranwal, V.K. Simultaneous detection of four garlic viruses by multiplex reverse transcription PCR and their distribution in Indian garlic accessions. J. Virol. Methods 2014, 202C, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Aloyce, R.C.; Tairo, F.; Sseruwagi, P.; Rey, M.E.C.; Ndunguru, J. A single-tube duplex and multiplex PCR for simultaneous detection of four cassava mosaic begomovirus species in cassava plants. J. Virol. Methods 2013, 189, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Yao, B.; Wang, G.; Ma, X.; Liu, W.; Tang, H.; Zhu, H.; Hong, N. Simultaneous detection and differentiation of three viruses in pear plants by a multiplex RT-PCR. J. Virol. Methods 2014, 196, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Usharani, T.R.; Laxmi, V.; Jalali, S.; Krishnareddy, M. Duplex PCR to detect both Papaya ring spot virus and Papaya leaf curl virus simultaneously from naturally infected papaya (Carica papaya L.). Indian J. Biotechnol. 2013, 12, 269–272. [Google Scholar] [CrossRef]
- Shen, W.; Tuo, D.; Yan, P.; Li, X.; Zhou, P. Detection of Papaya leaf distortion mosaic virus by reverse-transcription loop-mediated isothermal amplification. J. Virol. Methods 2014, 195, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Tuo, D.; Yan, P.; Yang, Y.; Li, X.; Zhou, P. Reverse transcription loop-mediated isothermal amplification assay for rapid detection of Papaya ringspot virus. J. Virol. Methods 2014, 204, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Sguazza, G.H.; Reynaldi, F.J.; Galosi, C.M.; Pecoraro, M.R. Simultaneous detection of bee viruses by multiplex PCR. J. Virol. Methods 2013, 194, 102–106. [Google Scholar] [CrossRef]
- Chamberlain, J.S.; Gibbs, R.A.; Ranier, J.E.; Nguyen, P.N.; Caskey, C.T. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 1988, 16, 11141–11156. [Google Scholar] [CrossRef] [PubMed]
- Noorani, M.S.; Awasthi, P.; Sharma, M.P.; Ram, R.; Zaidi, A.A.; Hallan, V. Simultaneous detection and identification of four cherry viruses by two step multiplex RT-PCR with an internal control of plant nad5 mRNA. J. Virol. Methods 2013, 193, 103–107. [Google Scholar] [CrossRef]
- Menzel, W.; Jelkmann, W.; Maiss, E. Detection of four apple viruses by multiplex RT-PCR assays with coamplification of plant mRNA as internal control. J. Virol. Methods 2002, 99, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Saade, M.; Aparicio, F.; Sanchez-Navarro, J.A.; Herranz, M.C.; Myrta, A.; Di Terlizzi, B.; Pallas, V. Simultaneous detection of the three ilarviruses affecting stone fruit trees by nonisotopic molecular hybridization and multiplex reverse-transcription polymerase chain reaction. Phytopathology 2000, 90, 1330–1336. [Google Scholar] [CrossRef]
- Thompson, J.; Wetzel, S.; Klerks, M.; Vašková, D.; Schoen, C.; Špak, J.; Jelkmann, W. Multiplex RT-PCR detection of four aphid-borne strawberry viruses in Fragaria spp. in combination with a plant mRNA specific internal control. J. Virol. Methods 2003, 111, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; Myrta, A.; Polak, J. Simultaneous detection and identification of four pome fruit viruses by one-tube pentaplex RT-PCR. J. Virol. Methods 2006, 133, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Lu, G.; Clover, G.R.G. A multiplex RT-PCR for the detection of Potato yellow vein virus, Tobacco rattle virus and Tomato infectious chlorosis virusin potato with a plant internal amplification control. Plant Pathol. 2009, 58, 203–209. [Google Scholar] [CrossRef]
- Matic, S.; Myrta, A.; Minafra, A. Detection of three closteroviruses in stone fruit trees by multiplex assays. J. Plant Pathol. 2010, 92, 57–63. [Google Scholar]
- GenBank, National Center for Biotechnology Information - NCBI. Available online: http://www.ncbi.nlm.nih.gov/genbank/ (accessed on 6 April 2014).
- Vector NTI Advance Software; Version 11.0; Invitrogen Corporation: Carlsbad, CA, USA, 2008.
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134–144. [Google Scholar] [CrossRef]
- BLASTN, National Center for Biotechnology Information - NCBI. Available online: http://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 10 May 2014).
- Yue, F.; Cui, S.; Zhang, C.; Yoon, K.J. A multiplex PCR for rapid and simultaneous detection of porcine circovirus type 2, porcine parvovirus, porcine pseudorabies virus, and porcine reproductive and respiratory syndrome virus in clinical specimens. Virus Genes 2009, 38, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Tecson Mendoza, E.M.; Laurena, A.C.; Botella, J.R. Recent advances in the development of transgenic papaya technology. Biotechnol. Annu. Rev. 2008, 14, 423–462. [Google Scholar] [PubMed]
- Fermin, G.A.; Castro, L.T.; Tennant, P.F. CP-Transgenic and non-transgenic approaches for the control of papaya ringspot current situation and challenges. Transgenic Plant J. 2010, 4, 1–15. [Google Scholar]
- Gonsalves, D. Control of Papaya ringspot virus in Papaya: A Case Study. Annu. Rev. Phytopathol. 1998, 36, 415–437. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Yang, L.; Liu, X.; Guan, X.; Jiang, L.; Zhang, D. Characterization of the exogenous insert and development of event-specific PCR detection methods for genetically modified Huanong No. 1 papaya. J. Agric. Food Chem. 2009, 57, 7205–7212. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Lu, G.; Clover, G. Novel approaches to mitigate primer interaction and eliminate inhibitors in multiplex PCR, demonstrated using an assay for detection of three strawberry viruses. J. Virol. Methods 2008, 151, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Shi, B.J.; Huang, X.G.; Peng, M.Y.; Zhang, X.M.; He, D.N.; Pang, R.; Zhou, B.; Chen, P.Y. A multiplex RT-PCR assay for rapid and differential diagnosis of four porcine diarrhea associated viruses in field samples from pig farms in East China from 2010 to 2012. J. Virol. Methods 2013, 194, 107–112. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tuo, D.; Shen, W.; Yang, Y.; Yan, P.; Li, X.; Zhou, P. Development and Validation of a Multiplex Reverse Transcription PCR Assay for Simultaneous Detection of Three Papaya Viruses. Viruses 2014, 6, 3893-3906. https://doi.org/10.3390/v6103893
Tuo D, Shen W, Yang Y, Yan P, Li X, Zhou P. Development and Validation of a Multiplex Reverse Transcription PCR Assay for Simultaneous Detection of Three Papaya Viruses. Viruses. 2014; 6(10):3893-3906. https://doi.org/10.3390/v6103893
Chicago/Turabian StyleTuo, Decai, Wentao Shen, Yong Yang, Pu Yan, Xiaoying Li, and Peng Zhou. 2014. "Development and Validation of a Multiplex Reverse Transcription PCR Assay for Simultaneous Detection of Three Papaya Viruses" Viruses 6, no. 10: 3893-3906. https://doi.org/10.3390/v6103893





