Host DNA Demethylation Induced by DNMT1 Inhibition Up-Regulates Antiviral OASL Protein during Influenza a Virus Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Viruses
2.2. Drug Treatment
2.3. Quantitative Real-Time PCR (qRT-PCR)
2.4. Plasmids, siRNAs and Other Reagents
2.5. Western Blot Analysis and Antibodies
2.6. DNMT Enzymatic Activity Assay
2.7. DNA Methylation Analysis
2.8. Immunofluorescence Staining
2.9. GO Annotation and KEGG Enrichment Analysis
2.10. Statistical Analysis
3. Results
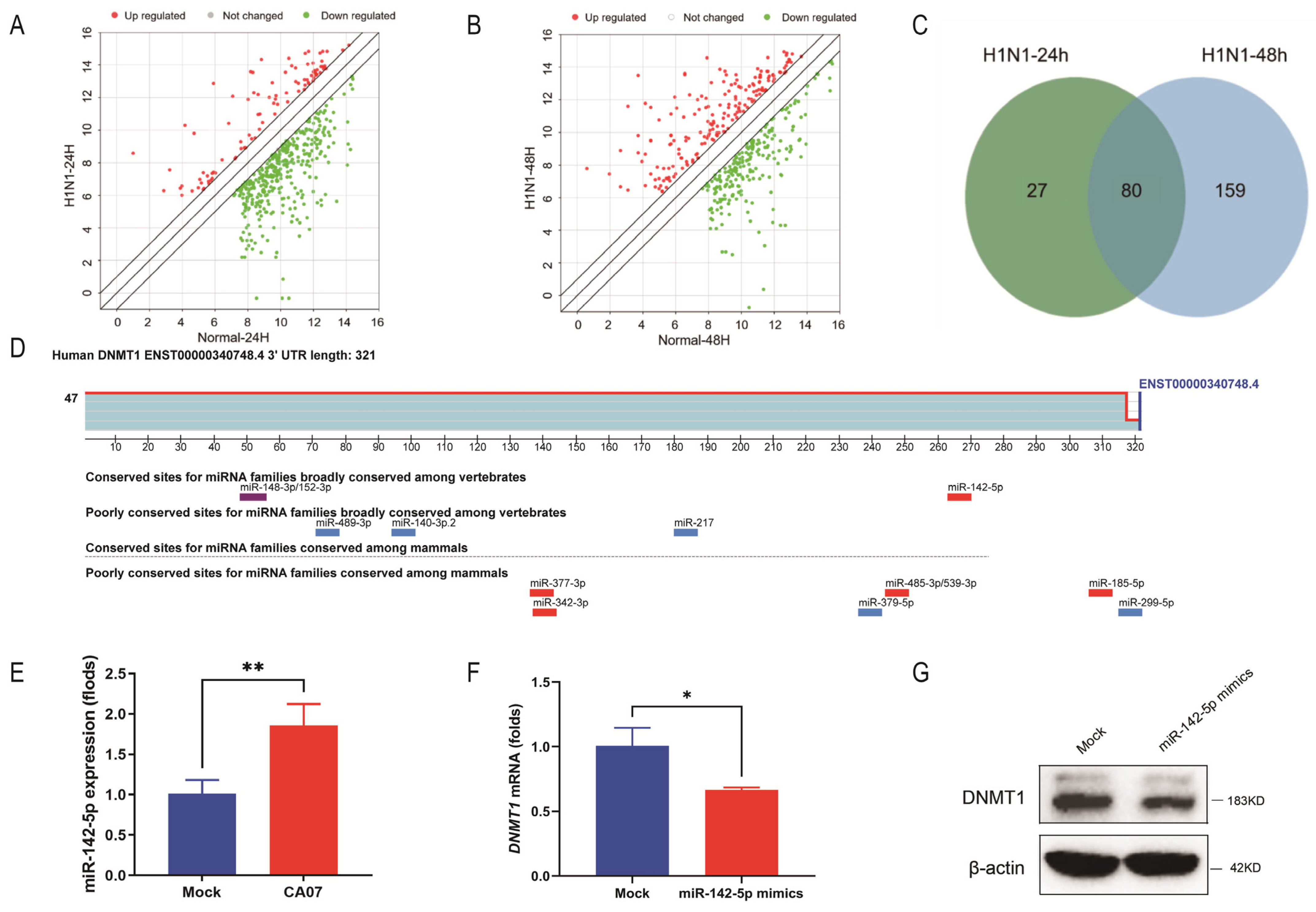
3.1. DNMT1 Expression Was Down-Regulated during IAV Infection
3.2. IAV Infection Inhibited DNMT1 Expression by Up-Regulating miR-142-5p
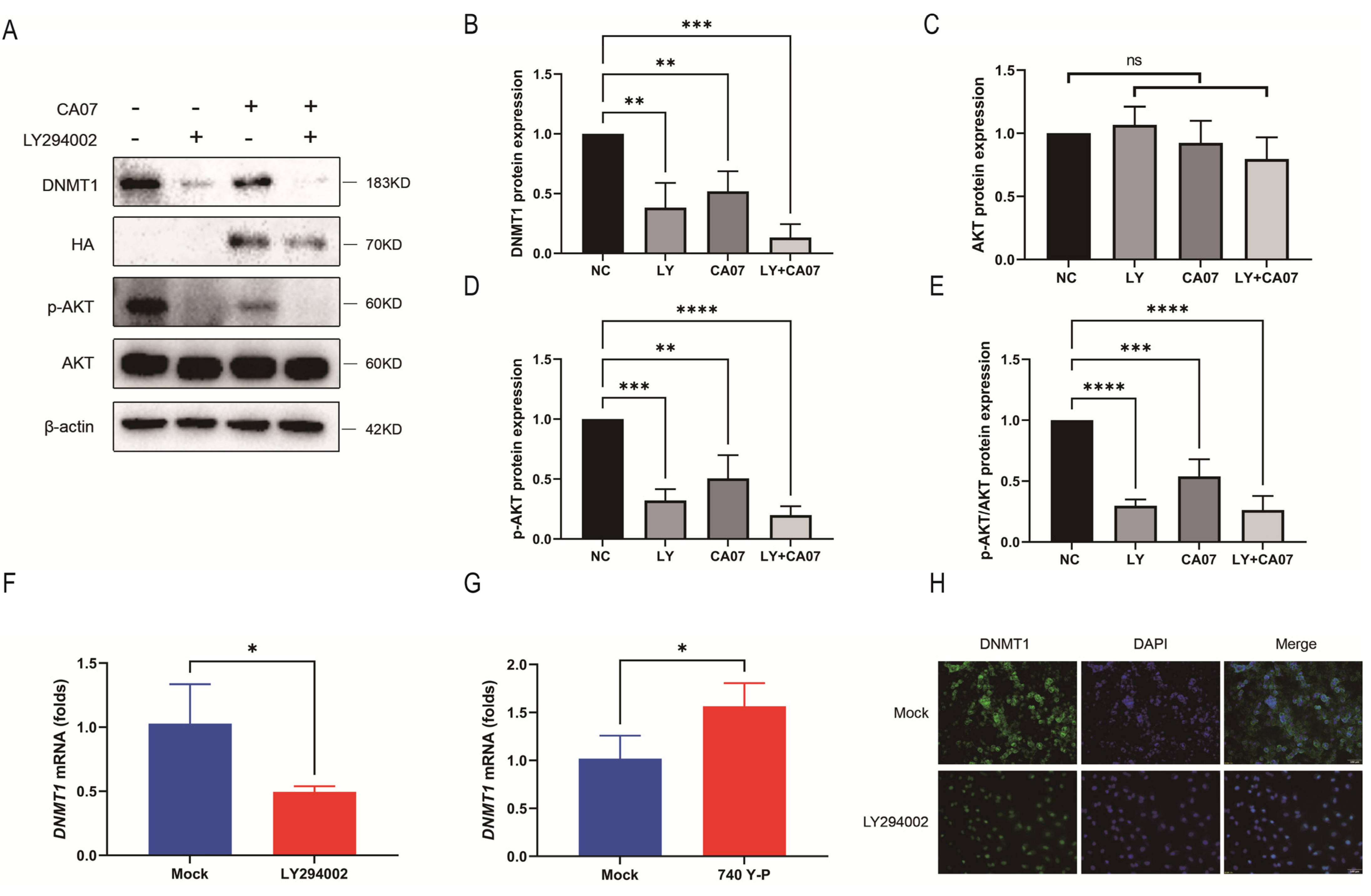
3.3. IAV Infection Inhibited the PI3K/AKT Signaling Pathway, Resulting in Down-Regulation of DNMT1
3.4. Oxford Nanopore Technologies Sequencing Screened Demethylated Genes
3.5. The Down-Regulated DNMT1 Expression Caused Demethylation of the OASL Promoter Region during IAV Infection
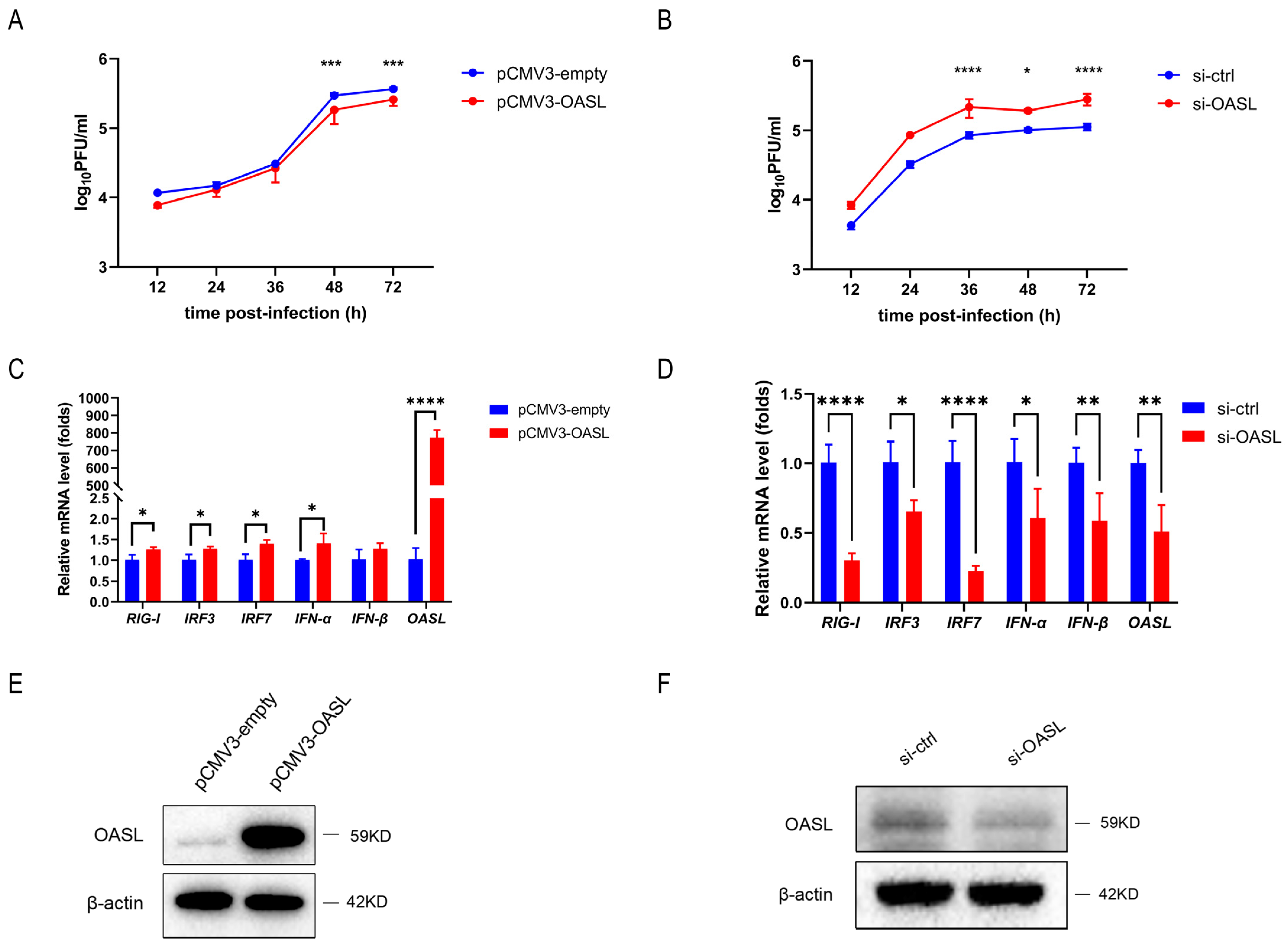
3.6. OASL Inhibited IAV Replication in A549 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ma, C.; Li, Y.; Zong, Y.; Velkov, T.; Wang, C.; Yang, X.; Zhang, M.; Jiang, Z.; Sun, H.; Tong, Q.; et al. p21 restricts influenza A virus by perturbing the viral polymerase complex and upregulating type I interferon signaling. PLoS Pathog. 2022, 18, e1010295. [Google Scholar] [CrossRef]
- Phan, T.; Fay, E.J.; Lee, Z.; Aron, S.; Hu, W.S.; Langlois, R.A. Segment-specific kinetics of mRNA, cRNA and vRNA accumulation during influenza infection. J. Virol. 2021, 95, 10–1128. [Google Scholar] [CrossRef]
- Sun, W.; Li, J.; Han, P.; Yang, Y.; Kang, X.; Li, Y.; Li, J.; Zhang, Y.; Wu, X.; Jiang, T.; et al. U4 at the 3′ UTR of PB1 segment of H5N1 influenza virus promotes RNA polymerase activity and contributes to viral pathogenicity. PLoS ONE 2014, 9, e93366. [Google Scholar] [CrossRef]
- Wang, P.; Song, W.; Mok, B.W.; Zheng, M.; Lau, S.Y.; Liu, S.; Chen, P.; Huang, X.; Liu, H.; Cremin, C.J.; et al. The PB2 Polymerase Host Adaptation Substitutions Prime Avian Indonesia Sub Clade 2.1 H5N1 Viruses for Infecting Humans. Viruses 2019, 11, 292. [Google Scholar] [CrossRef] [PubMed]
- Marcos-Villar, L.; Díaz-Colunga, J.; Sandoval, J.; Zamarreño, N.; Landeras-Bueno, S.; Esteller, M.; Falcón, A.; Nieto, A. Epigenetic control of influenza virus: Role of H3K79 methylation in interferon-induced antiviral response. Sci. Rep. 2018, 8, 1230. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Vipat, V.C.; Chakrabarti, A.K. Infection with influenza A viruses causes changes in promoter DNA methylation of inflammatory genes. Influenza Other Respir. Viruses 2013, 7, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Shao, L.; Sampath, P.; Zhao, B.; Patel, N.V.; Zhu, J.; Behl, B.; Parise, R.A.; Beumer, J.H.; O’Sullivan, R.J.; et al. Oligoadenylate-Synthetase-Family Protein OASL Inhibits Activity of the DNA Sensor cGAS during DNA Virus Infection to Limit Interferon Production. Immunity 2019, 50, 51–63.e55. [Google Scholar] [CrossRef] [PubMed]
- Ibsen, M.S.; Gad, H.H.; Andersen, L.L.; Hornung, V.; Julkunen, I.; Sarkar, S.N.; Hartmann, R. Structural and functional analysis reveals that human OASL binds dsRNA to enhance RIG-I signaling. Nucleic Acids Res. 2015, 43, 5236–5248. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, Y.; Ghosh, A.; Cuevas, R.A.; Forero, A.; Dhar, J.; Ibsen, M.S.; Schmid-Burgk, J.L.; Schmidt, T.; Ganapathiraju, M.K.; et al. Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor. Immunity 2014, 40, 936–948. [Google Scholar] [CrossRef]
- Leisching, G.; Ali, A.; Cole, V.; Baker, B. 2′-5′-Oligoadenylate synthetase-like protein inhibits intracellular M. tuberculosis replication and promotes proinflammatory cytokine secretion. Mol. Immunol. 2020, 118, 73–78. [Google Scholar] [CrossRef]
- Jones, M.J.; Goodman, S.J.; Kobor, M.S. DNA methylation and healthy human aging. Aging Cell 2015, 14, 924–932. [Google Scholar] [CrossRef]
- Uysal, F.; Cinar, O.; Can, A. Knockdown of Dnmt1 and Dnmt3a gene expression disrupts preimplantation embryo development through global DNA methylation. J. Assist. Reprod. Genet. 2021, 38, 3135–3144. [Google Scholar] [CrossRef] [PubMed]
- Mishima, Y.; Brueckner, L.; Takahashi, S.; Kawakami, T.; Otani, J.; Shinohara, A.; Takeshita, K.; Garvilles, R.G.; Watanabe, M.; Sakai, N.; et al. Enhanced processivity of Dnmt1 by monoubiquitinated histone H3. Genes Cells 2020, 25, 22–32. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Ko, M. Epigenetic Modification of Cytosines in Hematopoietic Differentiation and Malignant Transformation. Int. J. Mol. Sci. 2023, 24, 1727. [Google Scholar] [CrossRef]
- Dominguez, P.M.; Teater, M.; Chambwe, N.; Kormaksson, M.; Redmond, D.; Ishii, J.; Vuong, B.; Chaudhuri, J.; Melnick, A.; Vasanthakumar, A.; et al. DNA Methylation Dynamics of Germinal Center B Cells Are Mediated by AID. Cell Rep. 2015, 12, 2086–2098. [Google Scholar] [CrossRef]
- Zhou, S.J.; Deng, Y.L.; Liang, H.F.; Jaoude, J.C.; Liu, F.Y. Hepatitis B virus X protein promotes CREB-mediated activation of miR-3188 and Notch signaling in hepatocellular carcinoma. Cell Death Differ. 2017, 24, 1577–1587. [Google Scholar] [CrossRef] [PubMed]
- Hassouna, M.M.; Naguib, M.; Radwan, E.M.; Abdel-Samiee, M.; Estaphan, S.; Abdelsameea, E. DNA Methyltransferases as Potential Biomarkers for HCV Related Hepatocellular Carcinoma. Asian Pac. J. Cancer Prev. APJCP 2020, 21, 3357–3363. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Hong, L.; Cheng, C.; Li, N.; Zhao, X.; Shi, F.; Liu, J.; Fan, J.; Zhou, J.; Bode, A.M.; et al. DNMT1 mediates metabolic reprogramming induced by Epstein-Barr virus latent membrane protein 1 and reversed by grifolin in nasopharyngeal carcinoma. Cell Death Dis. 2018, 9, 619. [Google Scholar] [CrossRef]
- Tang, B.; Zhao, R.; Sun, Y.; Zhu, Y.; Zhong, J.; Zhao, G.; Zhu, N. Interleukin-6 expression was regulated by epigenetic mechanisms in response to influenza virus infection or dsRNA treatment. Mol. Immunol. 2011, 48, 1001–1008. [Google Scholar] [CrossRef]
- Jiménez-Wences, H.; Peralta-Zaragoza, O.; Fernández-Tilapa, G. Human papilloma virus, DNA methylation and microRNA expression in cervical cancer (Review). Oncol. Rep. 2014, 31, 2467–2476. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, J.; Li, J.; Yang, Y.; Kang, X.; Li, Y.; Wu, X.; Zhu, Q.; Zhou, Y.; Hu, Y. Up-regulation of microRNA-203 in influenza A virus infection inhibits viral replication by targeting DR1. Sci. Rep. 2018, 8, 6797. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Hao, Q.; Liu, L.; Li, Y.; Wu, J.; Huo, X.; Zhu, Y. Epigenetic changes mediated by microRNA miR29 activate cyclooxygenase 2 and lambda-1 interferon production during viral infection. J. Virol. 2012, 86, 1010–1020. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Sun, W.; Liu, L.; Yang, F.; Li, Y.; Chen, Y.; Fang, J.; Zhang, W.; Wu, J.; Zhu, Y. IL-32: A host proinflammatory factor against influenza viral replication is upregulated by aberrant epigenetic modifications during influenza A virus infection. J. Immunol. 2010, 185, 5056–5065. [Google Scholar] [CrossRef]
- Liu, S.; Liu, L.; Xu, G.; Cao, Z.; Wang, Q.; Li, S.; Peng, N.; Yin, J.; Yu, H.; Li, M.; et al. Epigenetic Modification Is Regulated by the Interaction of Influenza A Virus Nonstructural Protein 1 with the De Novo DNA Methyltransferase DNMT3B and Subsequent Transport to the Cytoplasm for K48-Linked Polyubiquitination. J. Virol. 2019, 93, 10–1128. [Google Scholar] [CrossRef]
- Zhang, X.; He, X.; Li, Q.; Kong, X.; Ou, Z.; Zhang, L.; Gong, Z.; Long, D.; Li, J.; Zhang, M.; et al. PI3K/AKT/mTOR Signaling Mediates Valproic Acid-Induced Neuronal Differentiation of Neural Stem Cells through Epigenetic Modifications. Stem Cell Rep. 2017, 8, 1256–1269. [Google Scholar] [CrossRef]
- Cisneros, F.J.; Branch, S. 5-AZA-2′-deoxycytidine (5-AZA-CdR): A demethylating agent affecting development and reproductive capacity. J. Appl. Toxicol. 2003, 23, 115–120. [Google Scholar] [CrossRef]
- Koehler, H.; Cotsmire, S.; Zhang, T.; Balachandran, S.; Upton, J.W.; Langland, J.; Kalman, D.; Jacobs, B.L.; Mocarski, E.S. Vaccinia virus E3 prevents sensing of Z-RNA to block ZBP1-dependent necroptosis. Cell Host Microbe 2021, 29, 1266–1276.e5. [Google Scholar] [CrossRef]
- Lee, S.A.; Chang, L.C.; Jung, W.; Bowman, J.W.; Kim, D.; Chen, W.; Foo, S.S.; Choi, Y.J.; Choi, U.Y.; Bowling, A.; et al. OASL phase condensation induces amyloid-like fibrillation of RIPK3 to promote virus-induced necroptosis. Nat. Cell Biol. 2023, 25, 92–107. [Google Scholar] [CrossRef]
- Gao, Z.J.; Li, W.P.; Mao, X.T.; Huang, T.; Wang, H.L.; Li, Y.N.; Liu, B.Q.; Zhong, J.Y.; Renjie, C.; Jin, J.; et al. Single-nucleotide methylation specifically represses type I interferon in antiviral innate immunity. J. Exp. Med. 2021, 218, e20201798. [Google Scholar] [CrossRef]
- Sardar, R.; Satish, D.; Gupta, D. Identification of Novel SARS-CoV-2 Drug Targets by Host MicroRNAs and Transcription Factors Co-regulatory Interaction Network Analysis. Front. Genet. 2020, 11, 571274. [Google Scholar] [CrossRef] [PubMed]
- Trobaugh, D.W.; Klimstra, W.B. MicroRNA Regulation of RNA Virus Replication and Pathogenesis. Trends Mol. Med. 2017, 23, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Liu, S.; Fabbri, M.; Liu, Z.; Heaphy, C.E.; Callegari, E.; Schwind, S.; Pang, J.; Yu, J.; Muthusamy, N.; et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood 2009, 113, 6411–6418. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, H.H.; Chen, Q.; Wang, Y.Y.; Fan, C.C.; Duan, Y.Y.; Huang, Y.; Zhang, H.M.; Li, J.P.; Zhang, X.Y.; et al. miR-142-5p Inhibits Cell Invasion and Migration by Targeting DNMT1 in Breast Cancer. Oncol. Res. 2022, 28, 885–897. [Google Scholar] [CrossRef]
- Schmolke, M.; Viemann, D.; Roth, J.; Ludwig, S. Essential impact of NF-kappaB signaling on the H5N1 influenza A virus-induced transcriptome. J. Immunol. 2009, 183, 5180–5189. [Google Scholar] [CrossRef]
- Ehrhardt, C.; Marjuki, H.; Wolff, T.; Nürnberg, B.; Planz, O.; Pleschka, S.; Ludwig, S. Bivalent role of the phosphatidylinositol-3-kinase (PI3K) during influenza virus infection and host cell defence. Cell. Microbiol. 2006, 8, 1336–1348. [Google Scholar] [CrossRef]
- Xu, B.; Peng, Y.J.; Zhu, W.J. Curcumin Inhibits Viability of Clear Cell Renal Cell Carcinoma by Down-Regulating ADAMTS18 Gene Methylation though NF-κ B and AKT Signaling Pathway. Chin. J. Integr. Med. 2022, 28, 419–424. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, S.; Yuan, Q.; Zhu, L.; Li, F.; Wang, H.; Kong, D.; Hao, J. TXNIP, a novel key factor to cause Schwann cell dysfunction in diabetic peripheral neuropathy, under the regulation of PI3K/Akt pathway inhibition-induced DNMT1 and DNMT3a overexpression. Cell Death Dis. 2021, 12, 642. [Google Scholar] [CrossRef]
- Sun, Y.; Xu, R.; Huang, J.; Yao, Y.; Pan, X.; Chen, Z.; Ma, G. Insulin-like growth factor-1-mediated regulation of miR-193a expression promotes the migration and proliferation of c-kit-positive mouse cardiac stem cells. Stem Cell Res. Ther. 2018, 9, 41. [Google Scholar] [CrossRef]
- Agarwal, S.; Amin, K.S.; Jagadeesh, S.; Baishay, G.; Rao, P.G.; Barua, N.C.; Bhattacharya, S.; Banerjee, P.P. Mahanine restores RASSF1A expression by down-regulating DNMT1 and DNMT3B in prostate cancer cells. Mol. Cancer 2013, 12, 99. [Google Scholar] [CrossRef]
- Blanco, J.; Cameirao, C.; López, M.C.; Muñoz-Barroso, I. Phosphatidylinositol-3-kinase-Akt pathway in negative-stranded RNA virus infection: A minireview. Arch. Virol. 2020, 165, 2165–2176. [Google Scholar] [CrossRef]
- Hirata, N.; Suizu, F.; Matsuda-Lennikov, M.; Edamura, T.; Bala, J.; Noguchi, M. Inhibition of Akt kinase activity suppresses entry and replication of influenza virus. Biochem. Biophys. Res. Commun. 2014, 450, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Ehrhardt, C.; Ludwig, S. A new player in a deadly game: Influenza viruses and the PI3K/Akt signalling pathway. Cell. Microbiol. 2009, 11, 863–871. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, C.; Shu, Y.; Ju, X.; Zou, Z.; Wang, H.; Rao, S.; Guo, F.; Liu, H.; Nan, W.; et al. Inhibition of autophagy ameliorates acute lung injury caused by avian influenza A H5N1 infection. Sci. Signal. 2012, 5, ra16. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Hui, J.; Mao, H. Nanopore Technology and Its Applications in Gene Sequencing. Biosensors 2021, 11, 214. [Google Scholar] [CrossRef]
- Liu, Y.; Rosikiewicz, W.; Pan, Z.; Jillette, N.; Wang, P.; Taghbalout, A.; Foox, J.; Mason, C.; Carroll, M.; Cheng, A.; et al. DNA methylation-calling tools for Oxford Nanopore sequencing: A survey and human epigenome-wide evaluation. Genome Biol. 2021, 22, 295. [Google Scholar] [CrossRef]
- Marques, J.; Anwar, J.; Eskildsen-Larsen, S.; Rebouillat, D.; Paludan, S.R.; Sen, G.; Williams, B.R.G.; Hartmann, R. The p59 oligoadenylate synthetase-like protein possesses antiviral activity that requires the C-terminal ubiquitin-like domain. J. Gen. Virol. 2008, 89, 2767–2772. [Google Scholar] [CrossRef]
- Dhar, J.; Cuevas, R.A.; Goswami, R.; Zhu, J.; Sarkar, S.N.; Barik, S. 2′-5′-Oligoadenylate Synthetase-Like Protein Inhibits Respiratory Syncytial Virus Replication and Is Targeted by the Viral Nonstructural Protein 1. J. Virol. 2015, 89, 10115–10119. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Z.; Li, J.; Feng, Y.; Kang, X.; Li, Y.; Chen, Y.; Li, W.; Yang, W.; Zhao, L.; Huang, S.; et al. Host DNA Demethylation Induced by DNMT1 Inhibition Up-Regulates Antiviral OASL Protein during Influenza a Virus Infection. Viruses 2023, 15, 1646. https://doi.org/10.3390/v15081646
Zhao Z, Li J, Feng Y, Kang X, Li Y, Chen Y, Li W, Yang W, Zhao L, Huang S, et al. Host DNA Demethylation Induced by DNMT1 Inhibition Up-Regulates Antiviral OASL Protein during Influenza a Virus Infection. Viruses. 2023; 15(8):1646. https://doi.org/10.3390/v15081646
Chicago/Turabian StyleZhao, Zhiyan, Jing Li, Ye Feng, Xiaoping Kang, Yuchang Li, Yuehong Chen, Wei Li, Wenguang Yang, Lu Zhao, Shenghai Huang, and et al. 2023. "Host DNA Demethylation Induced by DNMT1 Inhibition Up-Regulates Antiviral OASL Protein during Influenza a Virus Infection" Viruses 15, no. 8: 1646. https://doi.org/10.3390/v15081646
APA StyleZhao, Z., Li, J., Feng, Y., Kang, X., Li, Y., Chen, Y., Li, W., Yang, W., Zhao, L., Huang, S., Zhang, S., & Jiang, T. (2023). Host DNA Demethylation Induced by DNMT1 Inhibition Up-Regulates Antiviral OASL Protein during Influenza a Virus Infection. Viruses, 15(8), 1646. https://doi.org/10.3390/v15081646




